
AN EVOLUTIONARY APPROACH TO MULTIVARIATE FEATURE
SELECTION FOR FMRI PATTERN ANALYSIS
Malin
˚
Aberg, Line L
¨
oken and Johan Wessberg
Institute of Neuroscience and Physiology, G
¨
oteborg University, Box 432, SE-40530 Goteborg, Sweden
Keywords:
fMRI, pattern recognition, feature selection, evolutionary algorithms.
Abstract:
Multivariate pattern recognition has recently gained in popularity as an alternative to univariate fMRI ana-
lyis, although the exceedingly high spatial dimensionality has proven problematic. Addressing this issue, we
have explored the effectiveness of evolutionary algorithms in determining a limited number of voxels that,
in combination, optimally discriminate between single volumes of fMRI. Using a simple multiple linear re-
gression classifier in conjunction with as few as five evolutionarily selected voxels, a subject mean single trial
binary prediction rate of 74.3% was achieved on data generated by tactile stimulation of the arm compared
to rest. On the same data, feature selection based on statistical parametric mapping resulted in 63.8% correct
classification. Our evolutionary feature selection approach thus illustrates how, using appropriate multivariate
feature selection, surprising amounts of information can be extracted from very few voxels in single volumes
of fMRI. Moreover, the resulting voxel discrimination relevance maps (VDRMs) showed considerable overlap
with traditional statistical activation maps, providing a model-free alternative to statistical voxel activation
detection.
1 INTRODUCTION
We recently showed that the evolutionary algorithm
is an effective tool for classifier and feature subset
optimization for single-trial discrimination of electro-
encephalography (EEG) (
˚
Aberg and Wessberg, 2007).
In this study, we extend our approach to functional
magnetic resonance imaging (fMRI).
Similar to the EEG, fMRI data is non-stationary,
multivariate, noisy and very high-dimensional. These
properties are typically dealt with by applying statis-
tical parametric mapping (SPM) methods, where the
average level of voxel activity is computed offline in a
univariate, model-based fashion (Friston et al., 1994).
However, by being univariate, the SPM-based
method is not appropriately sensitive to cognitive in-
formation that is encoded in the combined effect of
numerous voxels. Pattern recognition approaches, on
the other hand, provide tools that are multivariate,
that is, based on the combined effect of several vox-
els. Moreover, trained pattern classifiers can be used
in situations that demand real-time results, including
online detection and identification of brain patterns.
Several recent studies have established the feasibility
of multivariate methods (Norman et al., 2006; Haynes
and Rees, 2006).
Due to the vast spatial dimensionality (in the or-
der of tens to hundreds of thousands of voxels), ef-
ficient feature selection has been identified as a ma-
jor challenge in the development of pattern classifica-
tion algorithms for fMRI (Norman et al., 2006). In
this study we therefore present an algorithm based on
evolutionary techniques, proven effective in numer-
ous optimization areas, including feature subset se-
lection (Hussein, 2001; Reeves and Rowe, 2002), that
detects which number and combination of individual
voxels that optimally carry information relevant to a
stimulus. These voxels are used as features in a classi-
fier, and we have chosen to use rudimentary multiple
linear regression (MLR) to show that even a very sim-
ple classification scheme can detect and distinguish
relevant cortical information in noisy fMRI data given
proper feature selection.
Our algorithm also generates a voxel selection
302
ËŽAberg M., Löken L. and Wessberg J. (2008).
AN EVOLUTIONARY APPROACH TO MULTIVARIATE FEATURE SELECTION FOR FMRI PATTERN ANALYSIS.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 302-307
DOI: 10.5220/0001064203020307
Copyright
c
SciTePress

frequency ranking, illustrating how relevant each
voxel is in discriminating between given patterns.
This ranking can be presented slicewise as a two-
dimensional image, or what we propose to call a voxel
discrimination relevance map (VDRM), showing the
anatomical location of brain regions involved in the
stimulus.
In this study we thus aim to evaluate the effective-
ness of the evolutionary approach in automatic voxel
subset selection, aspiring to improve single-volume
discrimination of cortical patterns. We also explore
how the results compare with established statistical
methods for detecting activated areas of the brain.
The data is acquired from a tactile stimulation exper-
iment where the physiology of brain activation is rea-
sonably well understood (Olausson et al., 2002). Part
of the findings have been previously presented in ab-
stract format (
˚
Aberg et al., 2006).
2 METHODS
Data Acquisition and Paradigm
A 1.5 T fMRI scanner (Philips Intera, Eindhoven,
Netherlands) with a sense head coil (acceleration fac-
tor 1) and a BOLD (blood oxygenation level depen-
dent) protocol with a T2*-weighted gradient echo-
planar imaging sequence (TR 3.5 s; TE 51 ms; flip an-
gle 90
◦
) was used to acquire brain scans in six healthy
human volunteers. The scanning planes (6 mm thick-
ness, 2.3 x 2.3 mm in-plane resolution) were oriented
parallel to the line between the anterior and posterior
commissure and covered the brain from the top of the
cortex to the base of the cerebellum. Each scan vol-
ume contained 25 slices at a spatial resolution of 128
x 128 voxels.
Following cues from the scanner, an experimenter
stroked a 7 cm wide soft brush over a 16 cm distance
in the distal direction on the right arm. Each brushing,
lasting 3.5 seconds (one single scan volume), was re-
peated three times and rest periods of equal duration
were interleaved. The Regional Ethical Review Board
at Goteborg University approved the study, and the
experiments were performed in accordance with the
Declaration of Helsinki.
Data Pre-processing
Data pre-processing was carried out with soft-
ware developed at the Montreal Neurologi-
cal Institute (Montreal, Canada; available at
http://www.bic.mni.mcgill.ca/software/). Functional
data were motion corrected and low-pass filtered with
a 6 mm full-width half-maximum Gaussian kernel.
Slices and voxels not containing brain matter were
discarded. To correct for hemodynamic delay, the re-
maining data (slices 2-20) was shifted by one volume.
An arm/rest data set containing 456 3.5 second pat-
terns of each class was formed per subject and slice,
and the samples were linearly normalized to the range
[0 1]. The first 80% of the patterns were randomized
and used in the evolutionary process (training data).
The remaining volumes were exclusively used in esti-
mating the prediction accuracy on already optimized
classifiers (validation data).
Feature Selection using Evolutionary Algorithms
An evolutionary algorithm is an optimization scheme
inspired by Darwinian evolution (Reeves and Rowe,
2002). The aim of the algorithm in this study is to se-
lect a limited number of voxels that, in combination
with a classifier, are maximally optimal in discrimi-
nating between the brain states induced by brushing
on the skin compared to rest.
Tournament selection is used here, where, for each
parent, a subset of individuals is randomly chosen
from the population and the fittest of these is selected.
The tournament size is set to a third of the total popu-
lation size. Reproduction is asexual, meaning that the
offspring is identical copies of the parents.
The fitness is computed as the proportion of cor-
rectly classified patterns using multiple linear regres-
sion. In order to avoid overfitting, the classifier pa-
rameters are established on the training data, whereas
a designated 25% of the training data (termed testing
data) is used for fitness computation.
The only mutation operation is substitution of a
voxel in the individual voxel subset with another, un-
used voxel. The frequency of mutation is regulated by
a constant mutation rate parameter.
Due to the stochastic nature of evolutionary algo-
rithms and the low signal-to-noise levels in the data,
the algorithm is unlikely to evolve the same voxel sub-
set at every attempt. To achieve robust results, the
algorithm is thus run numerous times.
The algorithm was implemented in Matlab (The
Mathworks, Massachusetts, USA) and C on a stan-
dard PC by one of the authors (M.
˚
Aberg).
Brain State Discrimination Performance
The prediction accuracy was evaluated on the valida-
tion data using the classifier and voxels from the run
that achieved best results on the training and testing
data. A discrimination accuracy of 50% corresponds
to chance.
AN EVOLUTIONARY APPROACH TO MULTIVARIATE FEATURE SELECTION FOR FMRI PATTERN
ANALYSIS
303

For comparison, the prediction accuracy using the
voxels with highest activation according to a statisti-
cal parametric mapping (SPM) method was also de-
termined. To this end, a statistical reference analysis
was performed on the training data (Worsley et al.,
2002).
Voxel Discrimination Relevance Maps
By aggregating the best feature subsets from each al-
gorithm run a voxel discrimination relevance ranking,
specifying the number of times each voxel has been
selected, can be obtained. This can be presented as a
slicewise two-dimensional voxel discrimination rele-
vance map (VDRM).
In order to mimic a classic block-design study
for comparison with the univariate SPM approach, all
data were used in the training process and there was
no prediction involved when generating the VDRMs.
3 RESULTS
Brain State Prediction Performance
The classification algorithm was applied to all sub-
jects individually using five voxels and 500 runs. The
prediction accuracies are well above chance (figure
1); a subject mean maximum prediction accuracy of
74.34% (range 65.79%-81.58%) was achieved. Us-
ing the five most active voxels as judged by the SPM
analysis, the subject mean maximum prediction ac-
curacy was significantly lower at 63.81% (Wilcoxon,
p=0.031; range 59.21-73.68%). Random classifica-
tion results in a prediction rate of 50%. Measuring the
prediction success in terms of information bits (Krip-
pendorf, 1986; Laubach et al., 2000), the difference
between methods is even more apparent: the SPMt-
based subject mean result is 0.094 bits (range 0.025-
0.22 bits), whereas the EA-based approach achieves
more than the double at 0.21 bits (range 0.077-0.32
bits).
The primary and secondary sensory cortices (SI
and SII), expected to be activated by tactile stimuli,
are approximately found in slices 4-7 and 11-12 (slice
numbers in the dorsal-ventral direction). As shown
in the bar chart in figure 2, the subject mean predic-
tion accuracies obtained in these slices are markedly
higher than in less relevant slices. Interestingly, the
prediction trend is clearly similar to the behavior of
the highest |SPMt|-value, a measure of brain activa-
tion (figure 2). It should be noted, however, that all
data is analyzed individually in native space rather
than at group level and that any anatomical congru-
ence between subjects is approximate at best.
50
60
70
80
1 2
3
4
5
6
|SPMt|-based
EA-based
prediction accuracy (%)
subject
Figure 1: Brain state prediction accuracies for all sub-
jects, as evaluated on the validation data set using the five
best voxels and corresponding classifiers obtained after 500
training runs. The prediction accuracy using the five most
activated voxels according to SPMt computations of the
training data is also shown. The level of chance is 50%.
slice number, dorsal-ventral direction –›
scaled measure
prediction accuracy
max |SPMt|-value
10
0
1
5 15 20
Figure 2: The subject mean brain state prediction perfor-
mance and maximum |SPMt|-values per slice. The two
variables show high correlation, and slices with voxels
where a BOLD response was expected (SI: slices 4-7, SII:
slices 11-12) show consistently higher values. The mea-
sures have been scaled to the range [0 1] within subjects to
emphasize trends.
Voxel Discrimination Relevance Maps
The VDRMs also show striking visual similarity to
the SPMt (figure 3), although the VDRMs appear less
noisy overall. SI (slices 5-6), for example, is detected
in the SPMt as well as in the VDRM. Similarly, the
location of SII (slices 11-12) and also the insular cor-
tex (slices 11), to which unmyelinated tactile afferents
project (Olausson et al., 2002), is clear from either
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
304
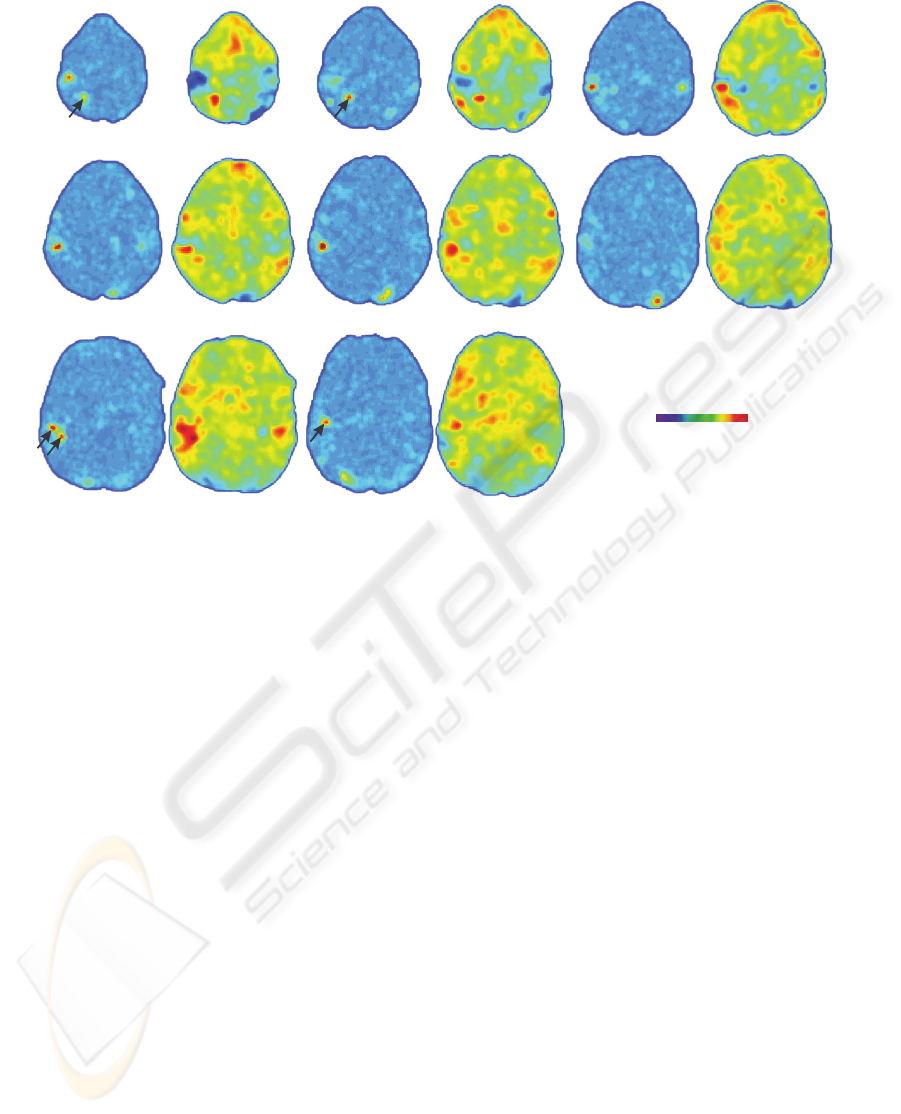
VDRM VDRM
SPM SPMSPM
6
8
10
5
7
9
VDRM
8.2-8.8
VDRM
SPMt
0
1
11 12
Figure 3: Voxel discrimination relevance maps for subject one, generated using five voxels over 500 runs, with corresponding
SPMts. The VDRMs and SPMts are clearly correlated. The right side of the images corresponds to the right side of the brain.
map. The SPMt also suggests several activated areas
that are not found in the VDRM. It should be noted,
however, that the SPMt maps are not thresholded, and
that all voxels with a |t|-value of less than 5.2 are
below the required significance levels. The VDRM
appears to detect highly activated negative and posi-
tive BOLD responses equally well, but does not dis-
tinguish between them (e.g. slice 5).
The Rffect of Voxel Subset Size
Including as few as two evolutionarily selected vox-
els yields voxel discrimination relevance maps where
some visual correlation with the SPM is clear (fig-
ure 4A). Further addition of voxels results in more
pronounced clustering at relevant sites, but also adds
noise. At 30 voxels the noise levels render the map
barely interpretable. Similarly, the subject mean
evolution-based prediction accuracy (figure 4B), in-
creases rapidly with the addition of up to three vox-
els, after which it levels out. Addition of more than 11
voxels decreases the performance drastically. SPMt-
based voxel selection behaves differently: the per-
formance for low numbers of voxels is poor, and in-
creases linearly with addition of voxels. Note that the
maximum number of available voxels is in the order
of thousands.
4 DISCUSSION
This study demonstrates the effectiveness of evolu-
tionary algorithms in selecting an optimal combina-
tion of voxels for highly accurate discrimination be-
tween single volumes of brain patterns — even in con-
junction with an exceedingly simple classifier. Us-
ing as few as five evolutionarily selected voxels and
a standard multiple linear regression classifier, a sub-
ject mean single-trial brain state prediction accuracy
of over 74% was achieved. Moreover, the voxel dis-
crimination relevance maps correlate clearly with sta-
tistical parametric maps, and the expected patterns
of brain activation were detected. Not surprisingly,
evolutionary feature selection achieved higher clas-
sification accuracy than voxel selection using SPM
ranking. The latter approach merely selects voxels
that show the largest individual average difference be-
tween brain states, whereas the evolutionary method
determines a combination of voxels that is tailored
for brain pattern discrimination. The feasibility of the
multivariate approach is further established by the fact
that the contribution of so little temporal and spatial
information — 3.5 seconds worth of data from only
five voxels — allows for accurate brain state predic-
tion. The maximum prediction accuracy using evolu-
tionary feature selection is achieved at drastically less
AN EVOLUTIONARY APPROACH TO MULTIVARIATE FEATURE SELECTION FOR FMRI PATTERN
ANALYSIS
305

2 voxels 4 voxels 10 voxels 18 voxels
SPMt
A
B
Number of voxels
1
0
Prediction
accuracy
1
2010
|SPMt|
EA
SPMt
-8.88.2
VDRM
0
1
Figure 4: A: The effect of number of included voxels on the voxel discrimination relevance maps for subject one, slice
five. The maximum number of possible voxels is in the order of thousands. B: Subject mean evolutionary and SPMt-based
prediction accuracy as a function of voxels subset size. The SPMt-based feature selection peaks at 65 voxels. The data has
been scaled within subjects to the range [0 1] to emphasize trends.
voxels than the SPMt approach (figure 4), indicating
that a large number of SPMt voxels are irrelevant for
the discrimination task. In addition, univariate fMRI-
analysis requires averaging over time to overcome the
inherently low signal quality, and lacks any prediction
qualities.
Our approach is not limited to brain state identi-
fication, but also provides two distinct approaches to
information localization. The fact that the slicewise
prediction accuracy correlates very well with the cor-
responding maximum |SPMt|-value — the classical
method of detecting activation — is a clear indication
that the information revealed by the prediction perfor-
mance is physiologically related to the stimulus (fig-
ure 2). The algorithm can be applied to voxel clusters
of any size and shape, defined either beforehand or
through evolution, thus optimizing the classification-
based information localization. Alternatively, the
voxel discrimination relevance maps serve as relative
activation detection charts, visually showing which
voxels are highly related to the stimulus. Signifi-
cance levels akin to SPMt values can be computed
using boot-strap statistical methods, involving data
permutations, allowing for proper VDRM threshold-
ing (Efron and Tibshirani, 1993). Although not done
here, the algorithm can be applied to a whole head
volume, resulting in a global rather than slicewise
VDRM.
In combination with excessive amounts of data,
typical for fMRI studies, the time taken to run an
evolutionary algorithm can be staggering. However,
in our design the number of included voxels is very
small, and using a standard PC (3.20GHz processor,
3GB RAM) one five-voxel training run on one indi-
vidual (20 slices) takes only approximately 1.5 min-
utes, whereas the validation is done in (biological)
real-time. Furthermore, several refinements can be
added to make the algorithm considerably more ef-
ficient.
The multiple linear regression method used for
classification in this study is sensitive to noise and
limited to linearly separable problems. In its sim-
plicity, however, the MLR effectively illustrates the
power of evolutionary algorithms in extracting rel-
evant information buried in substantial amounts of
noise. Pattern analysis using advanced non-linear al-
gorithms, such as artificial neural networks, have been
attempted and show promising results. Additional
discrimination algorithms, such as support vector ma-
chines and other state-of-the-art classifiers, can easily
be incorporated into the evolutionary scheme as re-
quired.
5 CONCLUSIONS
We have shown that evolutionary based multivoxel
feature selection is effective in extracting relevant
characterizing information from single volumes by
utilizing the multivariate properties of fMRI. More-
over, our approach provides a data-driven alternative
to voxel activation detection based on statistical meth-
ods.
ACKNOWLEDGEMENTS
This study was supported by the Swedish Research
Council (grant 3548), the Sahlgrenska University
Hospital (grant ALFGBG 3161), and the foundation
of Magnus Bergvall.
REFERENCES
˚
Aberg, M., L
¨
oken, L., and Wessberg, J. (2006). A multi-
variate pattern recognition approach to fMRI analysis
using linear classifiers combined with evolutionary al-
gorithms for voxel selection. Society for Neuroscience
36th Annual Meeting, Atlanta, USA, (492.11).
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
306

˚
Aberg, M. and Wessberg, J. (2007). Evolutionary op-
timization of classifiers and features for single-trial
eeg discrimination. BioMedical Engineering OnLine,
6(1):32.
Efron, B. and Tibshirani, R. (1993). An introduction to the
bootstrap. Chapman & Hall Ltd.
Friston, K. J., Holmes, A. P., Worsley, K. J., Poline, J. P.,
Frith, C. D., and Frackowiak, R. S. J. (1994). Statisti-
cal parametric maps in functional imaging: A general
linear approach. Human Brain Mapping, 2(4):189–
210.
Haynes, J.-D. and Rees, G. (2006). Decoding mental states
from brain activity in humans. Nature Reviews Neu-
roscience, 7(7):523–534.
Hussein, F. (2001). Genetic algorithms for feature selec-
tion and weighting, a review and study. In ICDAR
’01: Proceedings of the Sixth International Confer-
ence on Document Analysis and Recognition, page
1240, Washington, DC, USA. IEEE Computer Soci-
ety.
Krippendorf, K. (1986). Information theory: Structural
models for qualitative data. Sage University Paper
Series on Quantitative Applications in the Social Sci-
ences.
Laubach, M., Wessberg, J., and Nicolelis, M. (2000). Corti-
cal ensemble activity increasingly predicts behaviour
outcomes during learning of a motor task. Nature,
405:567–571.
Norman, K. A., Polyn, S. M., Detre, G. J., and Haxby, J. V.
(2006). Beyond mind-reading: multi-voxel pattern
analysis of fmri data. Trends in Cognitive Sciences,
10(9):424–430.
Olausson, H., Lamarre, Y., Backlund, H., Morin, C., Wallin,
B. G., Starck, G., Ekholm, S., Strigo, I., Worsley, K.,
Vallbo, A. B., and Bushnell, M. C. (2002). Unmyeli-
nated tactile afferents signal touch and project to insu-
lar cortex. Nature Neuroscience, 5(9):900–904.
Reeves, C. R. and Rowe, J. E. (2002). Genetic Algorithms
- Principles and Perspectives: A Guide to GA Theory.
Kluwer Academic Publishers, Norwell, MA, USA.
Worsley, K. J., Liao, C. H., Aston, J., Petre, V., Duncan,
G. H., Morales, F., and Evans, A. C. (2002). A gen-
eral statistical analysis for fmri data. NeuroImage,
15(1):1–15.
AN EVOLUTIONARY APPROACH TO MULTIVARIATE FEATURE SELECTION FOR FMRI PATTERN
ANALYSIS
307
