
PARSING TREE ADJOINING GRAMMARS USING
EVOLUTIONARY ALGORITHMS
Adrian Horia Dediu
GRLMC, Rovira i Virgili University, Pl. Imperial T`arraco 1, 43001, Tarragona, Spain
Faculty of Engineering in Foreign Languages, University “Politehnica” of Bucharest, Romania
C˘at˘alin Ionut¸ Tˆırn˘auc˘a
GRLMC, Rovira i Virgili University, Pl. Imperial T`arraco 1, 43001, Tarragona, Spain
Keywords:
Evolutionary Algorithms, Grammatical Evolution, Tree Adjoining Grammars, Parsing.
Abstract:
We use evolutionary algorithms to speed up a rather complex process, the tree adjoining grammars parsing.
This improvement is due due to a linear matching function which compares the fitness of different individuals.
Internally, derived trees are processed as tree-to-string representations. Moreover, we present some practical
results and a post running analysis that may encourage the use of evolutionary techniques in mildly context
sensitive language parsing, for example.
1 INTRODUCTION
Evolutionary algorithms (EAs) were introduced in
(Holland, 1962; Schwefel, 1965; Fogel, 1962), and
several were gradually developed during the past four
decades: evolutionary strategies (Rechenberg, 1973),
genetic algorithms (Holland, 1975) and evolutionary
programming (Fogel et al., 1966). Although they are
different approaches and independently studied, all
are inspired by the same principles and share common
components such as a searching space of individuals,
a coding scheme representing solutions for a given
problem, a fitness function or operators to produce
offspring. Roughly speaking, EAs try to stochasti-
cally solve searching problems by mimic of natural
principles of selecting and surviving the fittest indi-
vidual from a population. Real-world applications of
EAs deal with maximizing or minimizing objective
functions such as resource location or allocation opti-
mization.
Due to the lack of theoretical proofs, many for-
mal methods tried to model their behavior. For exam-
ple, EAs are simulated with the use of eco-grammar
systems in (Dediu and Grando, 2005). Networks of
evolutionary processors (NEPs) are another compu-
tational model having biological inspiration, and to-
gether with the above mentioned is closely related
to grammar systems (Castellanos et al., 2001). Al-
though NEPs are computationally complete (Csuhaj-
Varj´u et al., 2005), it is rather difficult for them to sim-
ulate EAs due to the difficulties given by the genera-
tion of the initial population with random individuals
and the implementation of a general fitness function
evaluation.
Recently, EAs have been also used for automatic
program generation such as LISP and the method of
genetic programming was established (Koza, 1992).
In (Ryan et al., 1998) even a more complex approach
which uses the parse trees of context-free grammars
and called grammatical evolution was proposed to
automatically evolve computer programs in arbitrary
languages.
Following this lead, we extended the applicabil-
ity of the parallel and cooperativesearching processes
of EAs to tree adjoining grammars parsing. We pro-
posed a new algorithm, called EATAG
P
, and we could
evolve derived trees using a tree-to-string representa-
tion (see Section 4.1). Implementing a linear match-
ing function to compare the yield of a derived tree
with a given input we obtained several encouraging
results during the running tests (see Section 4.2 and
Tables 2 and 1). For a better understanding of the al-
gorithm, a running example is given in Section 4.3.
Due to the high complexity of some classical pars-
ing algorithms, long sentences analysis could repre-
sent a very difficult task for a computer program. We
concluded that evolutionary algorithms used for pars-
ing process long sentences due to their reduced com-
632
Horia Dediu A. and Ionu¸t Tîrn
ˇ
auc
ˇ
a C. (2009).
PARSING TREE ADJOINING GRAMMARS USING EVOLUTIONARY ALGORITHMS.
In Proceedings of the International Conference on Agents and Artificial Intelligence, pages 632-639
DOI: 10.5220/0001811506320639
Copyright
c
SciTePress

putational complexity. In one of our examples, we
could implement a linear complexity fitness function.
Comparing with the O(n
6
) that is the complexity of
the classical parsing algorithm for TAGs, we could
increase the limit of the parsed words per sentence.
In the end of the paper we present a post run-
ning analysis (Table 2) that allowed us to propose sev-
eral research directions in order to extend the actual
known computational mechanisms in the mildly con-
text sensitive class of languages.
2 PRELIMINARIES
In this paper we follow standard definitions and nota-
tions in formal language theory. A wealth of further
information about this area can be found in (Hopcroft
and Ullman, 1990), and details about trees in (G´ecseg
and Steinby, 1997). An alphabet is a finite nonempty
set of symbols. A string is any sequence of symbols
from an alphabet X. The set of all strings over X is de-
noted by X
∗
, and subsets of X
∗
are called languages.
Moreover, |X| denotes the cardinality of the finite set
X, i.e., its number of elements, and N the set of non-
negative integers.
Now let us recall briefly the two core notions of
the present contribution: tree adjoining grammars
(TAGs) and evolutionary algorithms (EAs).
A TAG is a quintuple
T = (X,N, I,A,S) (1)
where:
• X is a finite alphabet of terminals,
• N is a finite alphabet of nonterminals,
• I is a finite set of finite trees called initial trees,
each of them having
– all interior nodes labeled by nonterminals, and
– all leaf nodes labeled by terminals, or by non-
terminals marked for substitution with ↓,
• A is a finite set of finite trees called auxiliary trees,
each of them having
– all interior nodes labeled by nonterminals, and
– all leaf nodes labeled by terminals, or by non-
terminals marked for substitution with ↓ except
for one node, called foot node, annotated by
“*”; the label of the foot node must be identical
to the label of the root node, and
• S ∈ N is a distinguished nonterminal called start
symbol.
A tree constructed from two other trees by us-
ing the two operations permitted in a TAG, substitu-
tion and adjoining, is called derived tree. Roughly
speaking, adjunction builds a new tree from an auxil-
iary tree and an initial, auxiliary or derived tree. On
the other hand, substitution replaces the node marked
with ↓ by the tree to be substituted (only trees de-
rived from the initial trees can be substituted for it).
For restrictions imposed on operations, examples and
details, the reader is referred to (Joshi and Schabes,
1997).
The tree set of a TAG contains all the trees that can
be derived from an S-rooted initial tree and in which
no node marked for substitution exists on the fron-
tier. The language generated by a TAG consists of
the yields of all trees in the tree set.
The other central notion, the EA, is defined as a
7-tuple
EA = (I, f,Ω,µ,λ,s,StopCondition) (2)
where:
• I is the set of the searching space instances called
individuals,
• f : I → F is a fitness function associated to indi-
viduals, with F a finite ordered set of values,
• Ω is a set of genetic operators (e.g., substitution,
mutation) which applied to the individuals of one
generation, called parents, produce new individu-
als called offspring,
• µ is the number of parents,
• λ is the number of offspring inside the population,
• s : I
µ
× I
λ
→ I
µ
is the selection operator which
changes the number of individuals from parents
and offspring producing the next generation of
parents (there are variants of EAs where after one
generation the parents are not needed anymore,
and in this case s selects only from the offspring,
i.e., s : I
λ
→ I
µ
), and
• StopCondition : F
µ
× N → {True,False} is the
stop criteria which may be interpreted as “Stop
when a good enough value was reached by an
individual fitness function”, “Stop after a certain
number of generations” or “Stop after a maximum
time available for computations”.
Note that sometimes associated with individuals we
can keep useful information for genetic operators.
It is usual to associate to each individual its fitness
value, using the notation h
−→
i
q
(u),Φ(
−→
i
q
(u))i, where
−→
i
q
(u) denotes the vectorial representation of the
chromosome of the q
th
individual in the generation
u and Φ(
−→
i
q
(u)) corresponds to the fitness value as-
sociated to that individual. Hence, we may consider
I = {h
−→
i
q
(u),Φ(
−→
i
q
(u))i | q,u ∈ N,1 ≤ q ≤ µ + λ}.
A good overview on EAs is (B¨ack, 1996).
PARSING TREE ADJOINING GRAMMARS USING EVOLUTIONARY ALGORITHMS
633

In Figure 1 we present a general description of
an EA, where gen is a numerical variable that indi-
cates the current number of generation, P(gen) rep-
resents the set of µ individuals that are potential par-
ents in generation gen and P’(gen) is the set of new
λ offspring that we get by the application of genetic
operators to individuals in P(gen). Depending on
the coding chosen for the EA each gene belongs to
a certain data type. Let L = (C
1
,C
2
,...,C
n
) be the
list of the sets of genes values, where n ∈ N is the
number of genes of the chromosomes. Without loss
of generality, we consider each C
j
, 1 ≤ j ≤ n, a fi-
nite and discrete set of values of a given type, such
that all values of genes in the position i of a chro-
mosome of an individual are of type C
i
. Also note
that in the Step 2 of the EA, (n,µ,L) initializes with
random values the chromosomes and is defined on
N × N ×C
1
×C
2
× .. . ×C
n
with values on I
µ
.
3 APPLYING EVOLUTIONARY
ALGORITHMS TO PARSE
CONTEXT-FREE GRAMMARS
Context-free grammars (CFGs) are a well-know class
of language generative devices extensively used for
describing syntax of programming languages and
some significant number of structures of natural lan-
guage sentences. For rigorous definitions and details,
the reader is referred to (Hopcroft and Ullman, 1990).
The tree structure of a string, called derivation tree,
shows how that string can be obtained by applying the
rules of the grammar. Note that a string can have mul-
tiple derivation trees associated. Parsing represents
the process of analyzing a string and constructing its
derivation tree. A parser is an algorithm that takes as
input a string and either accepts or rejects it (depend-
ing on whether it belongs or not to the language gen-
erated by the grammar), and in the case it is accepted,
also outputs its derivation trees. More about parsers
can be found in (Sippu and Soisalon-Soininen, 1988).
Due to the importance and applications of the
parsing algorithms, many efficient parsing techniques
were developed over the years. Yet there are sev-
eral computational models for which parsing is per-
formed in O(n
3
) for CFGs (Hopcroft and Ullman,
1990) or even O(n
6
) for more powerful generating
devices (Joshi and Schabes, 1997). For some long
sentences (e.g., more than 15 words), the computa-
tion time is rather large for practical approaches. In
this context, alternative parsing techniques appeared
in an attempt to reduce the complexity of the parsers,
sometime even with the price of inexactness (Schmid,
1997).
Grammatical Evolution (GE) is a new approach
proposedin (Ryan et al., 1998), which uses the deriva-
tion trees generated by CFGs and the searching capa-
bilities of EAs (especially genetic algorithms) to au-
tomatically evolve computer programs written in ar-
bitrary high-level programming languages. Such a
technique orders the productions for every nontermi-
nal of the CFG, and then uses the gene values to de-
cide which production to choose when it is necessary
to expand a given nonterminal. Recall that the genetic
coding (i.e., the sequence of chosen genes) is a string
of bytes.
Let us briefly describe the pioneering method of
(Ryan et al., 1998). Roughly speaking, EA finds a
function of one independent variable and one depen-
dent variable in symbolic form that fits a given sample
of 20 data points (x
i
,y
i
). The quadratic polynomial
function f(x) = x
4
+ x
3
+ x
2
+ x with values from the
interval [−1,1] was used with this purpose.
The considered CFG was:
1) hexpri ::= hexprihopihexpri
| (hexprihopihexpri)
| hpre− opi(hexpri)
| hvari
2) hopi ::= +| − |/|∗
3) hpre− opi ::= sin| cos|tan|log
4) hvari ::= X.
The algorithm constructs a symbolic expression
using a given sentential form. First, it starts with the
starting symbol hexpri, and it expands the leftmost
symbol considering the gene value modulo the num-
ber of choices (this way, the invalid gene value prob-
lem is solved). After that, the next leftmost symbol
is processed, and the next gene is used. If there is
only one choice, then the symbol is expanded without
considering the gene value. This procedure continues
until all the nonterminals in the sentential form were
expanded. If the string of genes is exhausted before
the nonterminals in the sentential form, then the string
of genes is used once again from the beginning as if
it were a circular string, so the problem of not having
enough genes is removed.
The fitness function evaluation promotes a multi-
criterial optimization, that is, maximizes the number
of fitting points and minimizes the error using the for-
mula
20
∑
i=1
| f(x
i
) − y
i
|. (3)
We can observe that GE cannot really evolve pro-
grams, only functions specified by samples.
ICAART 2009 - International Conference on Agents and Artificial Intelligence
634

The structure of an Evolutionary Algorithm is:
1 gen:=0;
2 Initialization process (n,µ,L);
3 Evaluate P(0):=
{h
−→
i
1
(0),Φ(
−→
i
1
(0))i,. .. ,h
−→
i
µ
(0),Φ(
−→
i
µ
(0))i};
4 do while not(StopCondition(Φ(
−→
i
1
(gen)),. .., Φ(
−→
i
µ
(gen)),gen))
5 Apply genetic operators;
6 Evaluate (P(gen))→ P
′
(gen)=
{h
−→
i
′
1
(gen),Φ(
−→
i
′
1
(gen))i,. ..,h
−→
i
′
λ
(gen),Φ(
−→
i
′
λ
(gen))i};
7 Select the next generation P(gen+1):=s(P(gen),P
′
(gen));
8 gen := gen+1;
end do;
Figure 1: The description of an evolutionary algorithm.
4 EVOLUTIONARY
ALGORITHMS FOR TREE
ADJOINING GRAMMARS
PARSING
Even if the usefulness of CFGs in representing
the syntax of natural languages is well established,
there are still several important linguistics aspects
that cannot be represented by this class of gram-
mars. For example, the languages multiple agreement
{a
n
1
a
n
2
...a
n
k
|n ≥ 1, k ≥ 3}, copy {ww|w ∈ {a,b}
∗
} and
cross agreement {a
n
b
m
c
n
d
m
|n,m ≥ 1} ⊂ {a,b,c,d}
∗
cannot be generated by any CFG. Thus, a more pow-
erful class of grammars, called tree adjoining gram-
mars (TAGs), was introduced (Joshi et al., 1975),
yielding interesting mathematical and computational
results over the past decades.
In this section we show how to apply a similar
technique with the one described in Section 3 to a
TAG in order to construct a derivation tree of a given
input string; progressively, we build at each step a de-
rived tree until we get a tree with the yield matching
the input string. This parsing algorithm for TAGs that
uses EAs will be called EATAG
P
in the following. Af-
ter presenting it, we point out some complexity issues
and show some tests performed.
Let us assume that for the rest of the paper the
TAG T = (Σ,N,I,A,S) and the input string is are
given.
4.1 The Algorithm EATAG
P
First we should mention that we adapted the tree-to-
string notation to simplify the internal representation
of the trees. We use curly brackets to specify the con-
straints, rectangular brackets to specify the children of
a node labeled by a nonterminal and a blank separator
after terminals. For nonterminals we have the con-
vention: they start with an uppercase letter followed
by other characters, then the specification of the con-
straints, and finally a “[” which marks the end of their
representation. Inside a balanced pair (“[”, “]”) we
have all the children of that nonterminal. A foot node
of an auxiliary tree has no children therefore there is
no need for the pair(“[”, “]”); instead, we have only
the foot marker “*”. An example is shown in the be-
ginning of Section 4.3 (see Figure 2 also).
The final goal of the algorithm is to find a derived
tree that has the root labeled with S and whose yield
matches the given input string is. To this end, we start
from an arbitrary S-rooted tree, and we may apply
substitutions and adjoinings to construct the target de-
rived tree. The searching process is exponential since
at every step several possible options to chose from
exist. From the beginning we may pick from several
S-rooted trees, then in the derived tree we may choose
from several nodes where to apply the next deriva-
tion, and once we do this, we may have several possi-
ble trees to substitute in or to adjoin at that particular
node.
EA’s individuals are viable solutions for the prob-
lem of the representation of a derived tree in a TAG
using a fixed number of genes, and they proved to
speed up this searching process. For the moment, let
us briefly explain how the algorithm works and which
are the issues that appear.
There are |I| initial trees and |I
S
| initial S-rooted
trees. We order all the trees in the sets I and A, respec-
tively, and all the nodes in every such tree according
to the node position in the tree-to-string representa-
tion. For example, if |I| = k, then we will identify
the trees in I by the numbers 1,2,. .., k. Thus, the
pairs (TreeNumber, NodeNumber) completely char-
acterize all the nodes in all the given trees of the TAG.
We start to construct a derived tree, and in this pro-
PARSING TREE ADJOINING GRAMMARS USING EVOLUTIONARY ALGORITHMS
635

gressive builded derived tree we carry on the nodes’
attributes such as substitution or adjoining constraints
(NA stands for non-adjoining).
We use the first gene modulo |I
S
| to select the start-
ing tree from the initial S-rooted trees, tree which will
transform into the desired derived tree after the fol-
lowing algorithm.
1. We repeat the algorithm’s steps until the length
of the yield of the derived tree will be greater or
equal than the length of the input string (stopping
condition). If this is not the case yet, and if we
have obligatory adjoining constraints, we should
satisfy them first.
2. Then we count how many nonterminals which do
not have the NA constraint are in the derived tree.
Let n
max
be this number. We use the next gene
modulo n
max
to select the next node, and there we
will apply a proper derivation. If somehow we
finish the genes, we start to use the string of genes
from the beginning.
3. Next, we proceed depending on the type of the
selected node:
(a) If the selected node is a substitution one, then
we count the trees that could be substituted in
our node, and let ns
max
be this number. We use
the next gene modulo ns
max
to select the next
substitution tree, and after that we perform the
substitution. Then, we go to Step 1.
(b) If the selected node is an adjoining node, then
we count the trees that could be adjoined at our
node, and let na
max
be this number. We use the
next gene modulo na
max
to select the next ad-
joining tree, and after that we do the adjoining.
Then, we go to Step 1.
Note that we can optimize the usage of genes, and
whenever we have a single option (node or tree) for
the next operation, we can perform the operation
without consuming the gene.
Now let us present the pseudocode for the infor-
mal description of the genetic decoding (a running
example can be found in Section 4.3). We mention
that in the line 17 of the algorithm, the “+” opera-
tor represents the usual concatenation for strings. We
describe only the adjoin part, the substitution being
similar (ng denotes the number of genes).
1) i=0 %counter for genes index
2) evolvedTree=initialTrees[gene[i] mod |I
S
|]
3) do while len(yield(evolvedTree)) < len(is)
4) i=(i+1) mod ng
5) n
max
=|internalNodes|-|internalNodes
with NA attributes|
6) adjNode=nonTerCandidates[gene[i] mod n
max
]
7) i=(i+1) mod ng
8) adjSet=adjNode.Label-type
9) na
max
=|adjSet|
10) i=(i+1) mod ng
11) insertedTree=adjSet[gene[i] mod na
max
]
12) t
1
=evolvedTree.substring(0,p
1
)
13) t
2
=insertedTree.split("*")[0]
14) t
3
=evolvedTree.substring(p
1
+1,p
2
)
15) t
4
=insertedTree.split("*")[1]
16) t
5
=evolvedTree.substring(p
2
)
17) evolvedTree=t
1
+t
2
+t
3
+t
4
+t
5
18) enddo
4.2 Fitness Function and Complexity
Fitness function assigns values to individuals devel-
oped by the EA, and that is why it is the most impor-
tant factor that directs the searching process. There-
fore, a fitness function that says “yes” or “no” to the
individuals of an EA is completely useless for the
searching process since the EA cannot know if a new
individual is a little bit better or worse than another
one.
In our algorithm we have to encourage two as-
pects: the matching of characters in the input string
and in the yield of the derived tree, and the equal
length of the two strings.
We can use several types of fitness function. For
example, the values associated to individuals are
triples of integers (k
1
,k
2
,k
3
), where k
1
represents the
maximum length of a sequence of matched charac-
ters, k
2
is the number of matches, and k
3
gets negative
values for yields longer than the input string. When
we make the comparisons between different individu-
als during the selection process, we consider the first
criterion the most important, then the second and then
the third.
Let M be the number of generations after which
we stop the evolution of the TAG. In our practical
implementation the crossover, mutation and selection
operators have the time complexity lower than the
fitness function evaluation complexity. Under these
circumstances, our algorithm EATAG
P
has the time
complexity O(M · (µ + λ) · Time( f)), where µ and
λ are as specified in the definition of an EA, and
Time( f) is the time complexity of the fitness func-
tion. During our tests we started with O(n
3
) time
complexity for the fitness function, then we reduced
it to O(n
2
), but the best results were obtained with a
linear fitness function.
4.3 Running Examples
For tests we used the TAG T = ({a},{S},{α
1
:
S{NA}[a S[a ]], α
2
: S{NA}[b S[b ]]}, {β
1
:
S{NA}[a S[S{NA} ∗ a ]], β
2
: S{NA}[b S[S{NA} ∗
b ]]},S) which generates the copy language {ww|w ∈
{a,b}
+
} (see Figure 2).
ICAART 2009 - International Conference on Agents and Artificial Intelligence
636
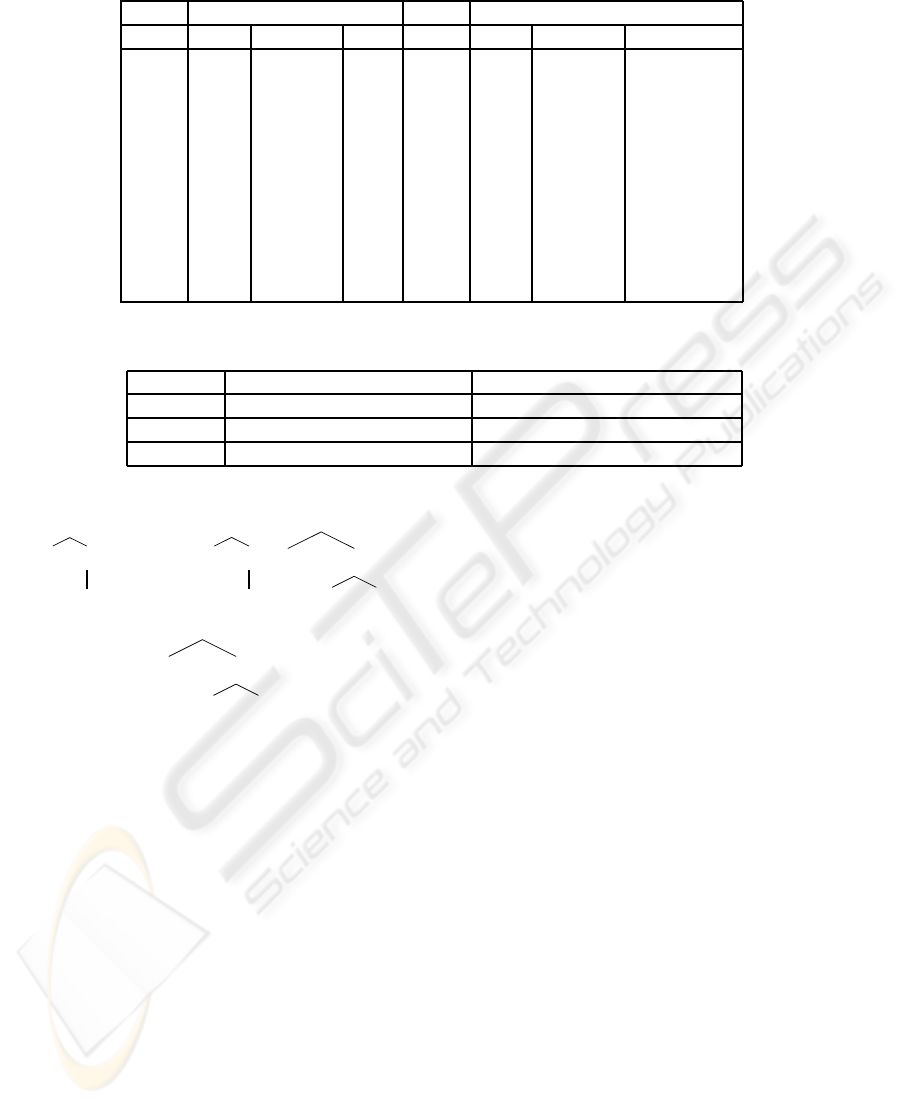
Table 1: Results of tests of EATAG
P
.
is: aaaabbbaaaaabbba is: aaaabbbaabbaaaaabbbaabba
GEN Max Average Min GEN Max Average Min
0 7 3.27 2 0 5 2.67 1
1 7 6.27 6 1 8 6.33 5
2 16 7.13 6 2 8 6.93 6
3 16 7.40 6 3 12 8.20 7
4 16 7.87 7 4 12 8.73 7
5 16 8.67 7 5 12 9.07 8
6 16 11.20 8 6 12 9.20 8
7 16 11.20 8 7 12 9.53 9
8 16 11.20 8 8 12 9.73 9
9 16 12.80 8 9 24 11.80 9
10 16 15.47 8 10 24 11.93 9
Table 2: Comparative tests for classical TAG parsing and EATAG
P
parsing.
First example len(input)=16 Second example len(input)=20
Computations Computations
classical 827,787 2,153,088
EATAG
P
268,886.3 661,745.5
α
1
:
S
NA
a S
a
α
2
:
S
NA
b S
b
β
1
:
S
NA
a S
S
NA
∗
a
β
2
:
S
NA
b S
S
NA
∗
b
Figure 2: The TAG T generating the copy language.
To explain better the decoding algorithm, we il-
lustrate it on an specific example. For the gram-
mar mentioned above and for the input string is =
“aaaabbbabbaaaabbbabb”, len(is) is 20, and the
number of initial S-rooted trees is 2. Moreover, we
have the following randomly generated sequence of
genes: 113,110,248,173,119,.. .. According to the
decoding algorithm’s line 2, gene[0] = 113, and the
evolvedTree is “S{NA} [b S[b ]]”. Since the length
of the yield of the evolvedTree is less than 20, the
algorithm continues with the while loop (code line
3). Next, n
max
is 1, and here due to the opti-
mization of genes’ usage we do not increment the
genes’ counter as described in code line 4. The
ad jNode position is 8 (strings’ index starts from
0), ad jNode.Label is “S”, na
max
is 2, gene[1]=248,
and the insertedTree is “S{NA}[a S[S{NA}*a
]]”. Moreover, we have p
1
= 8, p
2
= 13, t
1
=
“S{ NA}[b ”, t
2
= “S{NA}[a S[S{NA}”, t
3
= “[b ]”,
t
4
= “a ]]”, t
5
= “]”, and hence we get the evolvedTree
“S{ NA}[b S{NA} [a S[S{NA}[b ]a]]]”. The evolution
cycle continues generating more evolvedTree values
until we stop because the length of the yield of the
last derived evolvedtree (“baabbbaaaabaabbbaaaa”)
was 20.
Now that we explained how our algorithm works,
we turn our attention to the study of the behavior of
EATAG
P
(in terms of computations) for two input
strings, and then we present a comparison of the ob-
tained results with the ones from the classical parsing
algorithm for TAGs (Joshi and Schabes, 1997, p.102).
The tests were done for the input strings “aaaabb-
baaaaabbba” and “aaaabbbaabbaaaaabbbaabba” hav-
ing the lengths 16 and 24, respectively.
We used an EA with 15 individuals as the popula-
tion size, each having 20 genes with values between 0
and 255 (one byte). We considered the linear fitness
function as the maximum length of matching charac-
ters between the input string and the yield of the de-
rived tree.
Table 1 summarizes the results of the runs, where
GEN is the number of the generation, is represents
the input string, and Max, Average and Min, respec-
tively, are the best, the average and the worst, respec-
tively, fitness function’s values of individuals during
one generation.
Let us now compare EATAG
P
and the classical
TAG parsing. It is well known that theoretically, the
classical algorithm for parsing TAGs has the worst
case complexity O(n
6
). For many examples it does
PARSING TREE ADJOINING GRAMMARS USING EVOLUTIONARY ALGORITHMS
637

not reach the worst case, and we believe that the com-
parison of the results of the two parsing algorithms for
an average behavior will be more appropriate. On the
other hand, even if the EA uses a linear fitness func-
tion, the number of generations multiplied with the
number of individuals in the population could lead to
a significant volume of computations while solving
the parsing problem.
To make those comparisons, we used a rather em-
pirical method to measure the number of computa-
tions. In every cycle we incremented a global vari-
able called computations. We estimated the num-
ber of computations for both algorithms using the
same input string. One may argue that other com-
paring methods could be considered (e.g., measuring
the necessary time until finding the solution) but since
we made the implementations in two different pro-
gramming environments (VBA and Java), the running
time would have been influenced by other aspects, not
only by the complexity of the algorithms. By tak-
ing again the two input samples “aaaabbbaaaaabbba”
and “aaaabbbabbaaaabbbabb” having the length 16
and 20, respectively, we needed for the classical TAG
parsing algorithm only one run to determine the num-
ber of computations for each input example, while
for EATAG
P
we obtained the average result after ten
tests. All the results are synthesized in Table 2.
5 CONCLUSIONS AND FUTURE
WORK
We proposed an evolutionary algorithm for tree ad-
joining grammars’ parsing called EATAG
P
. After
some preliminary tests, we observed that the classi-
cal tree adjoining grammar parsing algorithm needs
approximatively three times more computations than
our algorithm to solve the same problem. Maybe it is
worthy to do more tests and further investigate under
what circumstances it performs better, if a conjecture
can be outlined or what improvements can be added.
We believe that our algorithm can be a turning point
in developing new models for knowledge base rep-
resentation systems or automatic text summarization,
for example.
As a drawback, for some examples EATAG
P
will
not be able to say that there is no solution. We could
have some doubts that we did not let the algorithm to
run enough generations, but in any case, we could run
tests for other examples, and we could approximate
the requested number of generations required to find
a solution for certain lengths of the input strings. This
will be done in the future.
Another intriguing aspect of EATAG
P
is that if the
grammar is ambiguous, we could find different pars-
ings for different individuals in the population during
one run for the same input string.
To conclude, we believe that our research could be
a starting point for developing new and more efficient
TAG parsing algorithms.
ACKNOWLEDGEMENTS
The work of A.H. Dediu was supported by Rovira i
Virgili University under the research program “Ra-
mon y Cajal” ref. 2002Cajal-BURV4. Many thanks to
the anonymous reviewers who encourage us and help
us improve the clarity and exposition of the present
material.
REFERENCES
B¨ack, T. (1996). Evolutionary algorithms in theory and
practice: evolution strategies, evolutionary program-
ming, genetic algorithms. Oxford University Press.
Castellanos, J., Mart´ın-Vide, C., Mitrana, V., and Sempere,
J. M. (2001). Solving np-complete problems with net-
works of evolutionary processors. In IWANN ’01, 6th
International Work-Conference on Artificial and Nat-
ural Neural Networks. Springer.
Csuhaj-Varj´u, E., Mart´ın-Vide, C., and Mitrana, V. (2005).
Hybrid networks of evolutionary processors are com-
putationally complete. Acta Inf., 41(4):257–272.
Dediu, A. H. and Grando, M. A. (2005). Simulating evo-
lutionary algorithms with eco-grammar systems. In
IWINAC’05, 1st International Work-conference on the
Interplay between Natural and Artificial Computa-
tion. Springer.
Fogel, L. (1962). Autonomous automata. Industrial Re-
search, 4:14–19.
Fogel, L. J., Owens, A. J., and Walsh, M. J. (1966). Arti-
ficial Intelligence through Simulated Evolution. The
University of Michigan Press, New York.
G´ecseg, F. and Steinby, M. (1997). Tree languages. In Sa-
lomaa, A. and Rozenberg, G., editors, Handbook of
formal languages, Vol. 3: beyond words, pages 1–68.
Springer, New York.
Holland, J. (1975). Adaptation in Natural and Artificial Sys-
tems. The University of Michigan Press.
Holland, J. H. (1962). Outline for a logical theory of adap-
tive systems. J. of the ACM, 9(3):297–314.
Hopcroft, J. E. and Ullman, J. D. (1990). Introduction
To Automata Theory, Languages, And Computation.
Addison-Wesley Longman Publishing Co., Inc.
Joshi, A. and Schabes, Y. (1997). Tree-adjoining grammars.
In Rozenberg, G. and Salomaa, A., editors, Handbook
of Formal Languages, Vol. 3: Beyond Words, pages
69–120. Springer, New York.
ICAART 2009 - International Conference on Agents and Artificial Intelligence
638

Joshi, A. K., Levy, L. S., and Takahashi, M. (1975). Tree
adjunct grammars. J. Comput. Syst. Sci., 10(1):136–
163.
Koza, J. (1992). Genetic Programming. The Theory of
Parsing, Translation, and Compiling. MIT Press.
Rechenberg, I. (1973). Evolutionsstrategie - Optimierung
technischer Systeme nach Prinzipien der biologischen
Evolution. Frommann-Holzboog, Stuttgart.
Ryan, C., Collins, J. J., and O’Neill, M. (1998). Gram-
matical evolution: Evolving programs for an arbitrary
language. In EuroGP’98, 1st European Workshop on
Genetic Programming. Springer.
Schmid, H. (1997). Parsing by successive approx-
imation. In IWPT’97, International Work-
shop on Parsing Technologies. Available
at
http://elib.uni-stuttgart.de/opus/
volltexte/1999/393/pdf/393_1.pdf
.
Schwefel, H.-P. (1965). Kybernetische Evolution als Strate-
gie der experimentellen Forschung in der Str¨omungs-
technik. Diplomarbeit, Technische Universit¨at Berlin,
Hermann F¨ottinger–Institut f¨ur Str¨omungstechnik.
Sippu, S. and Soisalon-Soininen, E. (1988). Parsing Theory
I: Languages and Parsing. Springer.
PARSING TREE ADJOINING GRAMMARS USING EVOLUTIONARY ALGORITHMS
639
