
INTERACTIVE 3D USER INTERFACES
FOR NEUROANATOMY EXPLORATION
Felix G. Hamza-Lup
1
and Tina Thompson
2
1
Computer Science, Armstrong Atlantic State University, Savannah, GA, U.S.A.
2
Biomedical Sciences, Mercer University, Macon, GA, U.S.A.
Keywords: User Interfaces, X3D, Interactive 3D, Neuroanatomy, e-Learning.
Abstract: Human neuroanatomy is extremely complex, and functional neuroanatomical pathways cannot be dissected
and easily visualized in an anatomy lab. Teaching students to see neuro-anatomical relationships over the
extent of the neuraxis is challenging. The ability to internalize a 3D map of the neuraxis with the appropriate
clinically relevant neuro-pathways superimposed is critical for medical students, as it facilitates long-term
retention of the information as opposed to short-term memorization. Interactive 3D simulations can play a
significant role in facilitating learning through engagement, immediate feedback, and by providing real-
world contexts.
1 INTRODUCTION
The human nervous system is the most complex
achievement of the process of evolution. It is the
primary mechanism in the detection of changes in
the external and internal environment triggering
appropriate responses in muscles, glands and organs.
Neuroanatomy is the study of the brain, spinal cord,
and peripheral nervous system. The understanding of
neuroanatomy correlation with function and
dysfunction is a cornerstone for future advances in
clinical neuroscience.
Medical students historically have difficulty
conceptualizing and projecting in their minds the 3D
aspects of the neuro pathways and embryonic organ
development from 2D text materials and electronic
resources. While highly specific laboratory tests and
sophisticated imaging techniques can be critical for
the practice of medicine, the basis of a neurological
exam relies on the physician’s ability to visualize
very complex neuroanatomical relationships, in
order to make highly accurate diagnosis.
Interactive web-based 3D simulations play a
significant role in facilitating learning through
engagement, immediate feedback, and providing
real-world contexts. An obvious benefit of
interactive systems with 3D models is the capability
to view spatial relationships among structures from a
variety of viewpoints.
The maturity of Web standards and
technologies has reached a point where development
and deployment of interactive 3D interfaces for
complex data visualization and knowledge sharing is
feasible.
In this paper we present our preliminary work
towards the development of an interactive Web
portal and a discussion on the usability paradigms
associated with this system. The project is in its
early stages of development and is yet to be
completed and evaluated. In Section 2 we provide a
brief overview of related work. In Section 3 we
present an interactive network that will simulate
nervous impulse propagation. In Section 4 we
discuss a few interaction design and assessment
issues. We close the paper with conclusions in
Section 5.
2 RELATED WORK
The practice of medicine relies on a clinician’s
ability to effectively integrate basic medical
knowledge with clinical experience to arrive at the
appropriate diagnosis. Laboratory tests as well as
sophisticated imaging techniques can supplement the
physician’s diagnostic skills. However, neurology is
one area of medicine which relies more on the
physician’s ability to use his/her knowledge of
130
Hamza-Lup F. and Thompson T.
INTERACTIVE 3D USER INTERFACES FOR NEUROANATOMY EXPLORATION.
DOI: 10.5220/0001823201300134
In Proceedings of the Fifth International Conference on Web Information Systems and Technologies (WEBIST 2009), page
ISBN: 978-989-8111-81-4
Copyright
c
2009 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
functional neuro-anatomical pathways to precisely
identify the cause and location of the underlying
medical problem (Adams et al, 2008).
Unfortunately, human neuro-anatomy is
extremely complex, and functional neuro-anatomical
pathways, while well described in books, cannot be
dissected and easily visualized in an anatomy lab.
Teaching students to “see” and understand neuro-
anatomical relationships over the extent of the
neuraxis (i.e., axial part of the central nervous
system) represents another level of complexity.
However, the ability to internalize a three-
dimensional map of the neuraxis with the
appropriate clinically relevant neuro-pathways
superimposed is critical for medical students, as it
facilitates long-term retention of the information as
opposed to short-term memorization (Mateen and
D’Eon, 2008). We hypothesize that the ability to
visualize neuro-anatomical pathways in 3D
significantly improves students’ ability to use
clinical deficits to localize discrete lesions.
While exploring the latest technology in 3D
content development for the Web, we followed the
VRML standard which pointed to eXtended 3D
(X3D). The Web3D consortium develops X3D as
an open standard for web-3D modeling and
information exchange. Within the Web3D
consortium, the MedX3D (Web3D, 2008) is tightly
focused on medical applications that can benefit
from real-time 3D visualizations. One of the
MedX3D research group’s focus is representation of
the human anatomy in X3D.
Another research effort to represent anatomy in
a Web setup is targeted at teaching anatomy
(Brenton et al., 2007). A major project, focused on
neuro-informatics tools for modeling the brain and
stressing the importance of modeling and sharing
data about the brain and its associated processes is
the Human Brain Project (Sheperd et al., 1998).
Our main goal is to develop an interactive
advanced learning system that will support
independent exploration and experimentation
through built-in features. The system will allow
students to manipulate in 3D and explore the
changes in the simulated process, and to visualize
motor and sensory systems as well as their
relationships over the neuraxis. At last but not least,
they will be able to visualize neural signal traveling
along the neuro-pathways from the triggered
receptor. The simulation could be presented as
supplementary material in the class, on the projector,
or as an assignment online, and will enhance and
complement the instructional material.
3 AN INTERACTIVE NETWORK
Primarily, two types of phenomena are involved in
processing nerve signals: electrical and chemical.
While electrical events propagate a signal within a
neuron, the chemical processes transmit the signal
from one neuron to another neuron or to a muscle
cell.
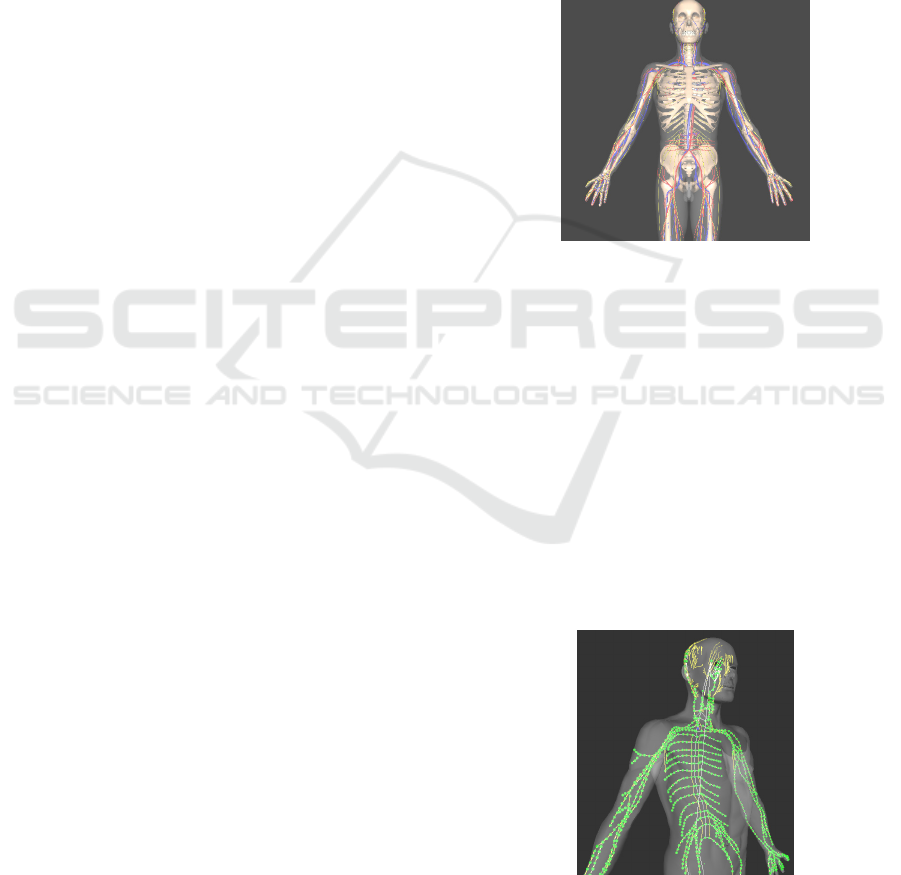
To visualize neural signal propagation along the
neuro-pathways, we developed an X3D anatomical
model based on a male dataset, as illustrated in
Figure 1.
Figure 1: An X3D anatomical male model.
The anatomical model consists of the following
subset: a skeletal system, a circulatory system, a
nervous system, a brain model, and the skin (outer
layer) model.
We decided to model the structure of the
nervous system as a directed graph with vertices that
follow the anatomical structures. The nodes
(vertices) in this graph are nervous excitation points
that can generate a nervous impulse. They
correspond to the triggered receptors of the nervous
system. Such a representation determines an
interactive network that can simulate nervous
impulse behavior at macroscopic and microscopic
levels.
Figure 2: Interactive Network (Macro Level). A graph
(tree) representation of the nervous system merged within
the X3D anatomical model.
INTERACTIVE 3D USER INTERFACES FOR NEUROANATOMY EXPLORATION
131
As the impulses travel along the nerves to the
neuraxis and the brain, they change color and shape.
The animation allows the user to visualize the path
taken by the impulses and the impulse behavior as it
crosses an anatomical structure. The X3D sensor-
based mechanism we implemented allows addition
and removal of nodes (i.e., trigger receptors) on the
skin of the 3D human model. The graph is editable,
and the student is able to develop its own network
and test its behavior.
Two modes are available: design mode and
simulation mode. In design mode, a student can load
a default network and continue to add location
sensors on the skin of the 3D model. By switching to
simulation mode the student can explore in 3D an
animation showing the propagation of the nervous
impulse throughout the nervous system.
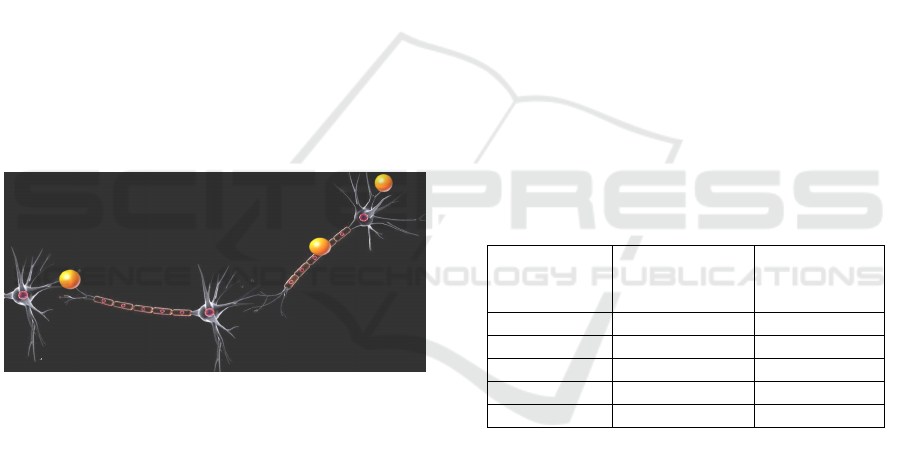
As the user zooms-in on the X3D network, after
a certain threshold is reached, the “Micro Level” is
activated and the chain of neurons associated with
that sub-section of the nervous system is rendered.
The microscopic level allows the user to visualize
the signal propagation at the microscopic level (as
illustrated in Figure 3, the signal represented as
yellow spheres that travel along the neurons chain).
Figure 3: Interactive Network (Micro Level).
The action potentials can travel along axons at
speeds of 0.1-100 m/s. This means that nerve
impulses can get from one part of a body to another
in a few milliseconds, which allows for fast
responses to stimuli. In the microscopic simulation,
the propagation speed of the signal is reduced to
provide a clear visualization of the neuron
components involved in the process. The level of
detail (LOD) change is currently under development.
To reduce the scene rendering time, we have
experimented with billboards with 2D neuron
textures mapped on them. The details of this module
will be presented in future articles.
4 INTERFACE INTERACTION
DESIGN
The goal of interaction design is to gain maximum
usability for our interface. In what follows we
explore several factors that will affect the system
interactivity and usability.
4.1 Optimization for Interactivity
In a 3D Web-based environment the scene graph has
to be loaded and cached on the client side. An X3D
file usually contains large datasets representing the
polygonal models in the scene. Linear
transformations are applied on the polygonal models
at loading time, most of the times. Such
transformations will slowdown the scene loading. To
speed up initial scene upload, we have investigated
compression algorithms for X3D. The X3D
representation of the male anatomical data set is
divided into the following subsets: skeletal system,
circulatory system, nervous system, the brain model,
and the skin. The compression algorithm reduces, on
average, three times the size of the model (as
illustrated in Table 1), and hence the X3D scene
loading time.
Table 1: Uncompressed and compressed X3D.
Body Part or
Subsystem
Before
Compression
(MB)
After
Compression
(MB)
Skeletal 3.04 1.01
Circulatory 12.41 3.81
Nervous 4.87 1.29
Brain 9.18 3.08
Skin 1.85 0.51
We are in the process of exploring compressed
binary encoding (ISO/IEC 19776-3, 2007) for the
X3D. Compressed binary encoding uses several
techniques to reduce the size of an X3D document
and to increase the speed of creating and processing
such documents. These techniques are primarily
based on the use of vocabulary tables that allow
small integer values to be used instead of character
strings.
Another optimization technique, based on the
initial position of the 3D models in the X3D scene,
is possible. For example, the male skin polygonal
model has currently around 18,000 polygons. As the
3D scene is initialized, a set of transformations will
be applied on each polygon in the model. This
computation done on-the-fly can significantly delay
WEBIST 2009 - 5th International Conference on Web Information Systems and Technologies
132

the loading time of the X3D scene at the client side.
We can pre-compute the transformations in the X3D
file and apply them to the “coordinate” sets
beforehand. We have implemented an optimizer
module that reduces the loading time in half by pre-
computing the transformations for each 3D object in
the scene.
4.2 Paradigms for Usability Support
Learnability is the ease with which new users can
begin effective interaction and achieve maximal
performance. Learnability is enhanced by several
paradigms like predictability, synthesizability, and
familiarity (Dix et al, 2004).
Predictability means that the user can easily
determine the results of his/her future actions on the
interface based on the interaction history. The X3D
interface is a consistent 3D environment that is fully
determined by the interaction history.
Synthesizability of the interface is very high since
the user is able to assess the effect of past operations
on the current state. One of the issues that may arise
is the X3D player’s robustness, i.e., parsing errors
may render parts of the scene invisible, having a
negative effect on predictability. In terms of
familiarity, the X3D interface navigation uses the
mouse buttons and their well-known functionality.
The 3D virtual objects have intrinsic properties that
suggest how they can be manipulated. Our informal
assessment shows that users familiar with the
window system have no difficulty in learning and
using the interface very fast. We have also deployed
a small size assessment experiment on a group of 12
students. The users were explained and asked to rank
the predictability of the system on a scale from 1 to
5 (1 meaning less predictable and 5 meaning very
predictable). The scores average was 4.83, denoting
a highly predictable system.
Another component for usability support is
flexibility. Flexibility represents the multiplicity of
ways the user and system exchange information.
Currently we are working at an interactive dialogue
system that will guide the user through various
simulations linked to a specific topic. We are also
investigating customizability and the transfer of
control for tasks execution, between the system and
the user, to support task migratability (i.e. the user
can have a computer assistant that will provide
guidance through certain parts of the simulation; the
simulation control could be switched from the user
to the computer at any time, to guide through
difficult sections). We are in the process of
developing an assessment experiment for the system
migratability in conjunction with the user task and
application domain.
5 CONCLUSIONS AND FUTURE
We have presented a few aspects of the early stages
of development of an advanced learning tool for
neuroanatomy. We have also discussed important
aspects of interactive interface design, as
interactivity is one of the main goals of the project.
Since there are different learning “speeds”, and
they vary from person to person, the learning tool is
available online in a Web-based environment
facilitating easy access anywhere and at anytime.
For neuroanatomy, theory is easier to grasp than to
translate into practice. In some cases, however,
practical skills are quickly achieved, even without
any basic understanding of the theory. In spite of
these difficulties, we want to achieve the best
theoretical and practical skills employing such
advanced learning tools.
We are currently developing a labeling system
that will allow students to visualize 3D neuro-
anatomical components and their associated names.
The labeling system will be accompanied by a
decomposition module. This module will allow
students to “virtually dissect” complex parts of the
central nervous system, as illustrated in Figure 4.
The figure denotes a 3D decomposition of the brain.
As the user “takes apart” the components of the
brain, s/he can better understand the location of the
parts within the system as well as the spatial
relationship among the nervous system components.
Figure 4: 3D Brain Model Decomposition.
A task of 3D model composition/decomposition
based on labeling will be used as a testing tool to
INTERACTIVE 3D USER INTERFACES FOR NEUROANATOMY EXPLORATION
133

assess student learning performance. We will report
our assessment results in future publications. The
tool is available online at www.neuro-pathways.org.
ACKNOWLEDGEMENTS
We would like to thank Mercer Medical School for
funding the initial development of the Neuro-
Pathways project and the student members of the
NEWS laboratory at the Armstrong Atlantic State
University. Website: www.neuro-pathways.org.
REFERENCES
Adams, M.E., Linn, J., and Yousry, I., 2008. “Pathology
of the ocular motor nerves III, IV, and VI”.
Neuroimaging Clinics of North America, 18 (2),
pp.261–282.
Brenton, H., Hernandez, J., Bello, F., Strutton, P.,
Purkayastha, S., Firth, T., and Darzi, A., 2007. “Using
multimedia and Web3D to enhance anatomy teaching”
Computers and Education, 49 (1), pp. 32–53.
Dix, A., Finlay, J., Abowd, G.D., Beale, R., 2004. “Part II
- Design Process, Chapter 7: Design rules” in Human-
Computer Interaction, Pearson Education Limited, 3
rd
edition, pp.258 – 288.
ISO/IEC 19776-3:2007 Information technology -
Computer graphics, image processing and
environmental data representation - Extensible 3D
(X3D) encodings - Part 3: Compressed binary
encoding.
Mateen, F., D’Eon, M., 2008. “Neuroanatomy: a single
institution study of knowledge loss”, Medical Teacher,
30 (5), pp 537–539.
Shepherd, G.M., Mirsky, J.S., Healy, M.D., Singer, M.S.,
Skoufos, E., Hines, M.S., Nadkarni, P.M., and Miller,
P.L.,1998. “The Human Brain Project:
neuroinformatics tools for integrating, searching and
modeling multidisciplinary neuroscience data”. Trends
Neuroscience, 21 (11), pp. 460–468.
Web3D, 2008. Medical real-time visualization,
communication using X3D. Last accessed Sept. 2008
from: www.web3d.org/x3d/workgroups/medical.
WEBIST 2009 - 5th International Conference on Web Information Systems and Technologies
134
