
DAMPING FACTOR CONSTRAINTS AND METABOLITE PROFILE
SELECTION INFLUENCE MAGNETIC RESONANCE
SPECTROSCOPY DATA QUANTIFICATION
M. I. Osorio Garcia, D. M. Sima, S. Van Huffel
Dept. Electrical Engineering, ESAT-SCD, Katholieke Universiteit Leuven, Kasteelpark Arenberg 10, 3001 Leuven, Belgium
F. U. Nielsen, U. Himmelreich
Biomedical Nuclear-Magnetic Resonance Unit, Katholieke Universiteit Leuven, Herestraat 49, 3000 Leuven, Belgium
Keywords:
Magnetic Resonance Spectroscopy (MRS), Quantification, Damping constraint.
Abstract:
Magnetic Resonance Spectroscopy (MRS) is a technique used for the diagnostics of tumour and metabolic
diseases by estimating the metabolite concentrations of the tissue under investigation. Unreliable metabolite
estimation may mislead the diagnosis and therefore quantification of MRS in vivo signals must be performed
carefully. In this work, we quantify 1.5 Tesla (T) and 9.4 T MRS in vivo signals and study the influence of the
damping factor constraint and the metabolite profile selection used in the quantification method. The damping
factor bounds the linewidth of the metabolite profiles and may yield bad fits if wrongly selected. Furthermore,
MRS data quantification leads to overestimation of some metabolite concentrations when the selected metabo-
lite basis set is incomplete suggesting that metabolites are fitting the region of their neighboring components.
Here, we evaluate the normality of the residual which in cases of good fitting contains no metabolites and
only white Gaussian noise. Furthermore, we propose to estimate the damping bound adaptively by taking into
account information from the linewidth of the signal and the metabolite basis set.
1 INTRODUCTION
Magnetic Resonance Spectroscopy (MRS) is a non-
invasive technique used to estimate the metabolite
concentration of living tissue. MRS is used in the di-
agnosis of cancer, epilepsy, metabolic and other dis-
eases because it provides information about the bio-
chemical condition of a tissue. Acquisition is per-
formed in the time domain, resulting in Free Induction
Decay (FID) signals, and the conversion into the fre-
quency domain using the Fourier transform is called
the MR spectrum. A variety of quantification meth-
ods exist for estimating the metabolite concentrations
using either the time or the frequency domain data
(Ratiney et al., 2004; Provencher, 2001; Poullet et al.,
2007). For quantifying MRS signals we make use of
the time domain method presented in (Poullet et al.,
2007), where a basis set of reference metabolites is
employed for estimating the metabolite concentra-
tions with the model in Eq.(1). In the ideal case when
the metabolite basis set completely describes the sig-
nal under investigation and the noise on the signal is
white, the method in (Poullet et al., 2007) is a max-
imum likelihood approach. A thorough investigation
of the MRS noise and the conditions for this noise be-
ing white complex Gaussian are presented in (Grage
and Akke, 2003). The model that describes the MRS
signals under investigation is:
K
∑
k=1
a
k
e
( jφ
k
)
e
(−d
k
t+2πj f
k
t)
v
k
(t) + B(t) (1)
where K is the number of metabolites, j =
√
−1, v
k
(t)
the given metabolite profile k in the basis set, a
k
the
unknown amplitude, φ
k
the unknown phase shift cor-
rection, d
k
the unknowndamping, f
k
the unknownfre-
quency shift and B(t) is the baseline. In this case we
also measured in vivo the macromolecular contribu-
tion which is included in the basis set and decreases
the contribution of B(t). Among these parameters,
the most essential are the amplitudes a
k
, since they
are proportional to the concentration. The quantifica-
tion method described by (Poullet et al., 2007) esti-
mates the unknown parameters a
k
, d
k
, f
k
and φ
k
using
176
I. Osorio Garcia M., M. Sima D., Van Huffel S., U. Nielsen F. and Himmelreich U..
DAMPING FACTOR CONSTRAINTS AND METABOLITE PROFILE SELECTION INFLUENCE MAGNETIC RESONANCE SPECTROSCOPY DATA
QUANTIFICATION.
DOI: 10.5220/0003139301760181
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2011), pages 176-181
ISBN: 978-989-8425-35-5
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
a nonlinear least squares problem for fitting model (1)
to the MRS signal. To encourage a reliable and mean-
ingful fit, extra constraints, such as equal phases and
some bounds on the frequencies and dampings are
imposed on these parameters. Quantification is com-
monly evaluated visually by checking the residual and
numerically by checking the Cram´er-Rao bounds of
the fitted metabolites. A fit can be considered un-
successful when metabolites are not well-estimated
(large Cram´er-Rao bounds) or the residual contains
metabolite contributions. Graphical statistical mea-
sures for residual analysis are useful together with nu-
merical measures because they are directly related to
visual inspection of the entire data set at once and can
easily point out a range of relationships between the
model and the data. On the other hand, numerical
measures are more focused on a particular property
of the data and often try to compress that information
into a single number. Depending on the data and anal-
ysis requirements, one might need to use both types
of measures to evaluate the quality of the fit. A well-
fitted signal contains no metabolites in its residual and
therefore the residual should contain only white com-
plex Gaussian noise.
In this paper, we focus on two aspects that have
strong influence on the quantification method’s per-
formance. First, we show that mis-specification of
the damping factor constraint in the quantification
method of (Poullet et al., 2007) is systematically re-
flected in the residual of the fit. Second, we exam-
ine the influence of the number and importance of the
reference metabolites used in the basis set and how
residual analysis can help in identifying incomplete
metabolite basis sets. To evaluate the goodness of
the fit we compute a quality factor proposed by (Slot-
boom et al., 2009) and we extend it to estimate prob-
lematic frequency regions in the residual by using a
moving window. Furthermore, we employ the nor-
mal probability plot, the cumulative probability and
the Rayleigh distribution to study the behavior of the
complex residual. In particular we assess whether it
contains only Gaussian noise or whether systematic
patterns from the metabolites are still present.
2 MRS SIGNALS
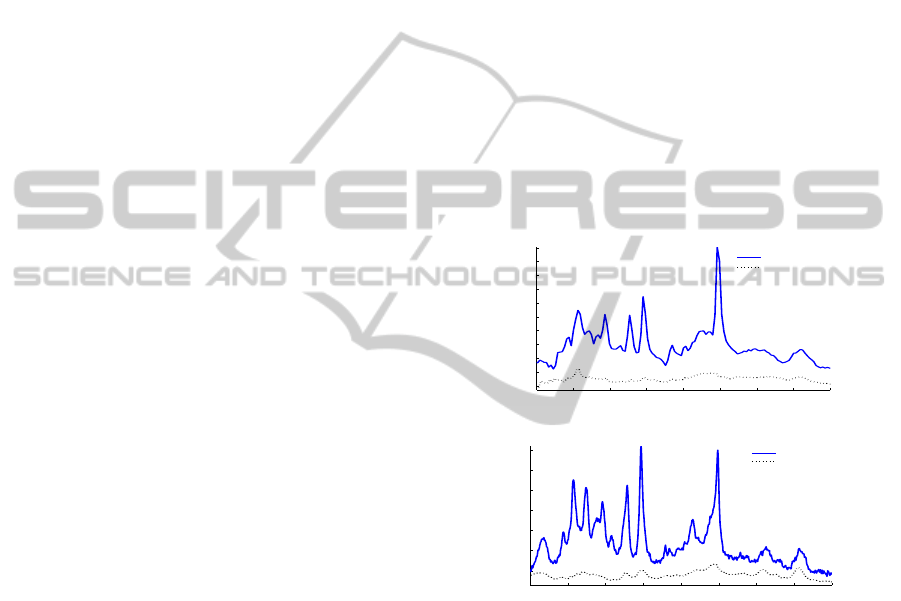
We analyze two types of signals:
• An
1
H MRS signal from human brain acquired
at 1.5 Tesla (T) on a Philips NT Gyroscan scan-
ner. This signal was obtained using the PRESS
pulse sequence (Bottomley, 1984). MRS parame-
ters were: repetition time of 6s, TE = 23 ms, SW
= 1 KHz and 64 averages. B0 eddy current correc-
tion (Klose, 1990) was performed using the water
reference before quantification. See Fig.1 (a).
• An
1
H MRS signal from rat brain acquired at
9.4 T on a Bruker Biospec small animal MR scan-
ner (Bruker BioSpin MRI, Ettlingen, Germany).
This signal was acquired using the PRESS pulse
sequence (Bottomley, 1984) with implemented
pre-delay Outer Volume Suppression (OVS) and
making use of the water suppression method VA-
POR (Tk´aˇc et al., 1999). MRS parameters were:
repetition time of 8s, TE = 20 ms, SW = 4 KHz
and 128 averages. B0 eddy current correction as
well as B0 drift removalwere performed using the
Bruker built-in routines and shimming was per-
formed using FASTMAP (Gruetter, 1993). See
Fig.1 (b).
Additionally, an unsuppressed water signal is always
measured, which is commonly used as a reference for
phase and lineshape corrections.
0.511.522.533.544.5
0
0.02
0.04
0.06
0.08
0.1
0.12
0.14
0.16
0.18
0.2
ppm
Amplitude [a.u.]
Signal 1.5T
MM
(a)
0.511.522.533.544.5
0
2
4
6
8
10
12
x 10
5
ppm
Amplitude [a.u.]
Signal 9.4T
MM
(b)
Figure 1: Real part of the in vivo spectra. The dotted curve
is the nulled metabolite signal corresponding to the mea-
sured macromolecules and lipids. (a) Human brain signal
acquired at 1.5T. (b) Rat brain signal acquired at 9.4T.
On both scanners, an in vitro basis set of reference
metabolites was measured. The following metabo-
lites were used: Alanine (Ala), Aspartate (Asp), Crea-
tine (Cre), Gamma-Aminoburytic acid (GABA), Glu-
cose (Glc), Glutamine (Gln), Glutamate (Glu), Glyc-
erolphosphorylcholine (GPC), Glutathione (GSH),
Lactate (Lac), Myo-Inositol (m-Ins), N-Acetyl As-
partate (NAA), Phosphorylcholine (PCh), Phospocre-
atine (PCr), Phosphoryl Ethanolamine (PE) and Tau-
rine (Tau). A typical problem for in vivo MRS quan-
tification is the presence of a macromolecular sig-
nal affecting the baseline of the spectra. For this
DAMPING FACTOR CONSTRAINTS AND METABOLITE PROFILE SELECTION INFLUENCE MAGNETIC
RESONANCE SPECTROSCOPY DATA QUANTIFICATION
177

study, we measured the in vivo spectrum of macro-
molecules (MM) using an inversion recovery se-
quence. The inversion time was fine-tuned experi-
mentally for metabolite nulling and this MM signal
was added to the basis set of reference metabolites.
3 QUANTIFICATION
3.1 Preprocessing
In in vivo
1
H MRS signals, the concentration of water
in the brain is several orders of magnitude higher than
the concentration of the metabolites. This signal is
suppressed during acquisition in order to increase the
resolution of the metabolites of interest. Nevertheless,
there is always some residual water around 4.7 ppm
that must be removed to reduce the complexity of the
signal analysis and improve the quantification accu-
racy. The signals presented here were filtered using
a method called Hankel Singular Value Decomposi-
tion (Pijnappel et al., 1992), which decomposes the
FID in a sum of exponentials and eliminates the com-
ponents found in the specific frequency region of the
water. In particular, the more efficient HLSVD-PRO
implementation (Laudadio et al., 2002) is used. Other
preprocessing steps for these signals include phase
correction for better visualization of the spectra, and
time circular shift for the Bruker signals, which were
performed using the jMRUI software package (Stefan
et al., 2009).
3.2 AQSES
The quantification method used here for analyzing the
spectra is AQSES (Poullet et al., 2007). This is a time-
domain method that combines metabolite profiles in
the best way as expressed in Eq.(1) to fit the signal
under analysis. The most important output parame-
ters of this method are the amplitudes of each metabo-
lite, because these are proportional to the concentra-
tion of metabolites in the tissue. However, the quan-
tification method also requires the tunning of other
model parameters such as small frequency shifts f
k
,
damping corrections d
k
and a common phase term
φ
k
=φ for each metabolite. In particular, the damp-
ing parameters allow the metabolite profiles to be nar-
rower or wider for better fitting the signal. An es-
sential model parameter is the upper bound for the
damping parameters used as a constraint in the quan-
tification method AQSES. This is needed in order to
avoid metabolite profiles to become too broad and
fit the baseline. However, a too low upper bound
is not desired, as metabolites are then badly fitted.
Until now this damping bound was chosen as a fix
value independent on the signal information. In this
study, we propose to estimate this bound as an adap-
tive method that takes into account information from
the signal and the metabolite basis set. To this end, we
make use of the fact that the damping of a complex
damped exponential is equal to the linewidth (i.e. the
Full Width at Half Maximum (FWHM)) of the corre-
sponding Lorentzian peak. Thus, we approximate the
upper bound for the damping factor constraint as the
difference between the FWHM of the unsuppressed
water signal of the in vivo and a singlet from the in
vitro metabolites (i.e. NAA). As can be seen in Fig.1,
there is a macromolecular background signal underly-
ing the in vivo signal. Baseline correction in AQSES
is accounted for by simultaneously using the MM sig-
nal in the basis set as well as a smooth spline model
for additional baseline correction.
3.3 Statistical Residual Analysis
The residual is the difference between the measured
signal and the fit of this signal as obtained by AQSES.
We use the residual in the frequency domain either
for graphical assessment of the goodness-of-fit, or as
an indication of possible fitting problems. If the fit
is correct, the residual should not contain metabolites
and should be white noise. To assess the goodness of
the fit, we use a quality factor proposed by (Slotboom
et al., 2009): Q
fit
(N) =
R
2
N.σ
2
, where σ
2
is the variance
of the signal noise calculated from the metabolite-free
region, R is the norm of the residual and N is the num-
ber of points of the least-squares fit. This value of Q
fit
is close to 1 when the fit is perfect, bigger than 1 when
the model is probably incomplete (lack of metabo-
lites) and smaller than 1 when parts of the noise are
fitted (overfitting) which means that the model has too
many degrees of freedom.
4 RESULTS AND DISCUSSION
4.1 Effect of Damping Factor
Constraint
We study the effect of the damping factor constraint,
which allows each metabolite in the basis set to be as
narrow or wide as the in vivo signal. Figure 2 shows
the results of quantification for 3 different damping
factor constraint values. Quantification can be first
of all evaluated by visual inspection of the residual,
which already provides information about the regions
that are not well-fitted. Setting the upper bound on the
BIOSIGNALS 2011 - International Conference on Bio-inspired Systems and Signal Processing
178
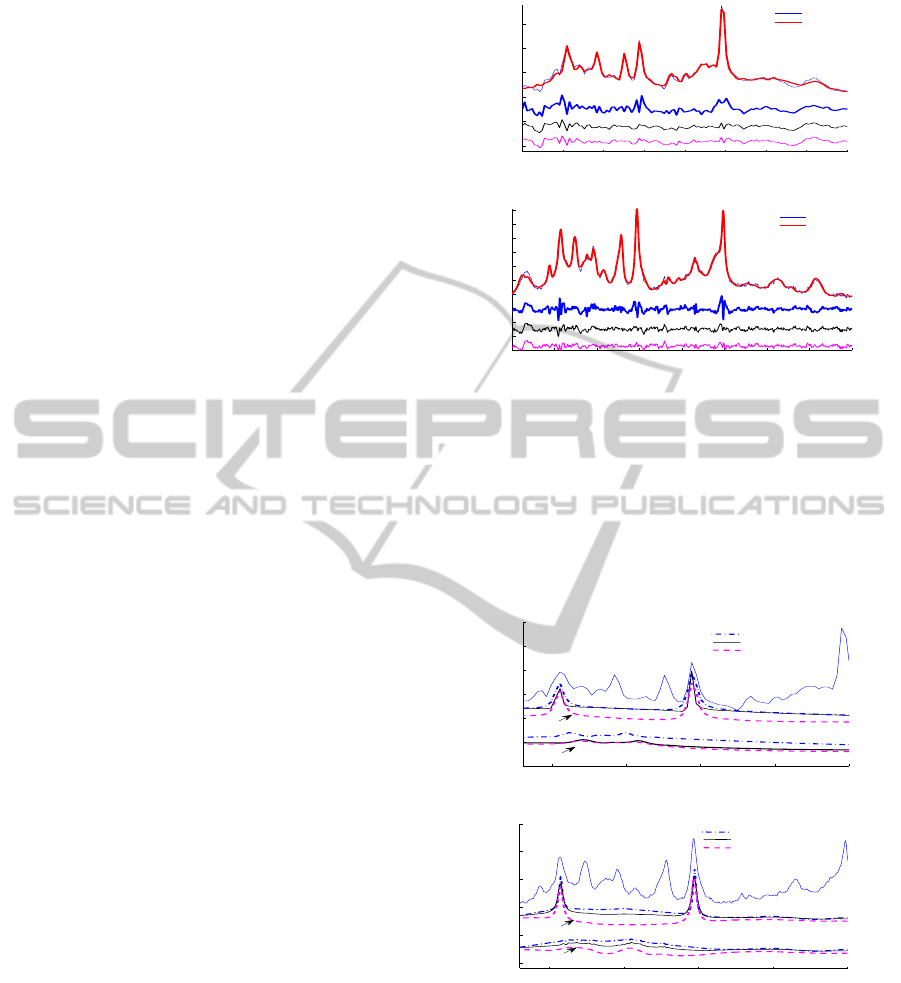
damping factors too low is clearly reflected in the pat-
tern of the residual, where peaks with similar shape
are observed in the region of the metabolites. Subse-
quently, when a bigger damping is allowed, improve-
ments in the fitting are reflected in the residual (see
Fig.2, e.g. around 2 ppm, 3 ppm and 3.9 ppm). Al-
though large increases of the damping with regards to
the good bound do not improve or worsen the resid-
ual, a more detailed look into the results illustrate that
when a too large damping factor is allowed, some
metabolites may wrongly take over other metabolites
or baseline contribution (see especially the bottom
profile of Glc in Fig.3). Moreover, this effect is not
obvious from the fitting results and it may affect the
stability of the method, leading to wrong estimations.
Correlation is also used to evaluate the goodness
of the fit by correlating the original signal with the fit-
ting. Values close to 1 reflect a good correlation and
thus a good fit. However, a good correlation does not
mean good estimates and therefore we must carefully
interpret this parameter. The correlations of the orig-
inal and the fitted signal for the three damping val-
ues at 1.5 T were 0.9234, 0.9770 and 0.9786 respec-
tively; and the correlations for the 9.4 T signal were
0.9241, 0.9808 and 0.9861 respectively. For testing
the assumption that the residual estimated after a good
fit is random noise (i.e. white Gaussian noise), we
evaluated and tested the real and imaginary parts of
the residuals. A zero mean bivariate normal variable
Z = (X,Y) with X andY uncorrelated with equal vari-
ances, σ
2
X
= σ
2
Y
= σ can be expressed in polar coor-
dinates as Z = (Rcos(θ), Rsin(θ)) where the radius
R has a Rayleigh distribution with scale parameter σ
and the angle θ is uniformly distributed on the interval
[−π, π] (Grage and Akke, 2003). This assumption has
been used to assess whether the real and imaginary
parts of the residual are independent and identically
distributed (results not shown here).
4.2 Lack of Metabolites in the Basis Set
Additionally, we study the effect of the number of
metabolites used for quantification as this may also
cause over- or underestimation. Fig.4 shows the re-
sults of amplitude estimation for the 1.5 T and 9.4 T
signals when using a complete and incomplete basis
set of metabolites. For the 1.5 T signal we considered
3 groups of metabolite profiles: (a) all 13 metabo-
lites (Ala, Asp, Cho, Cre, GABA, Glc, Gln, Glu, Lac,
Myo, NAA, Tau), (b) the 6 most relevant metabolites
having a high concentration in normal brain (Cho,
Cre, Gln, Glu, Myo, NAA), (c) the 4 most relevant
metabolites having the highest concentration (Cho,
Cre, Glu, NAA). For the 9.4 T signal we also con-
0.511.522.533.544.5
−0.1
−0.05
0
0.05
0.1
0.15
ppm
Amplitude [a.u.]
Signal
Fit
(1) Residual damping 0.001
(2) Residual damping 0.01
(3) Residual damping 0.5
(a)
0.511.522.533.544.5
−8
−6
−4
−2
0
2
4
6
8
10
12
x 10
5
ppm
Amplitude [a.u.]
Signal
Fit
(1) Residual damping 0.0025
(2) Residual damping 0.05
(3) Residual damping 0.5
(b)
Figure 2: Quantification results using different damping
factor constraints which reflect the over- or underestima-
tion of amplitude estimates and the importance of its care-
ful selection. The best fitted signal is the overlapped thick
line and the residuals are the curves beneath. (a) 1.5 T sig-
nal and (b) 9.4 T signal. (1) Residual with damping factor
constraint 0.001 (small bound). (2) Residual with damp-
ing 0.01 factor constraint (good bound). (3) Residual with
damping 0.5 factor constraint (big bound).
22.533.54
−0.1
−0.05
0
0.05
0.1
0.15
0.2
ppm
Amplitude [a.u.]
Glc
Cre
Small bound
Good bound
Big bound
(a)
22.533.54
−1
−0.5
0
0.5
1
1.5
x 10
6
ppm
Amplitude [a.u.]
Small bound
Glc
Cre
Good bound
Big bound
(b)
Figure 3: Effect of damping factor constraint in metabolites
shows a higher impact in small metabolites. The curves be-
neath the signal correspond to metabolite estimates of Crea-
tine and Glucose using small, good and big damping factor
constraints. (a) 1.5 T signal and (b) 9.4 T signal.
sidered 3 groups of metabolite profiles: (a) all 16
metabolites (Ala, Asp, Cre, GABA, GPC, GSH, Glc,
Gln, Glu, Lac, Myo, NAA, PCh, PCr, PE, Tau), (b)
the 8 more relevant metabolites having a high concen-
tration (Cre, GPC, GSH, Gln, Glu, Myo, NAA, PE),
DAMPING FACTOR CONSTRAINTS AND METABOLITE PROFILE SELECTION INFLUENCE MAGNETIC
RESONANCE SPECTROSCOPY DATA QUANTIFICATION
179

Ala Asp Cho Cre GABA Glc Gln Glu Lac Myo NAA Tau
0
0.5
1
1.5
2
Amplitude / Cre
Literature (Provencher)
Estimates 12 Met
Estimates 6 Met
Estimates 4 Met
(a)
Ala Asp Cre GABAGPC GSH Glc Gln Glu Lac Myo NAA PE Tau
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
1.8
2
Amplitude / Cre
Literature (Cudalbu)
Literature (Pfeuffer)
Estimates 16 Met
Estimates 9 Met
Estimates 4 Met
(b)
Figure 4: Amplitude estimates and comparison using a
complete and incomplete basis set. (a) Amplitude estima-
tion of metabolites relative to Cre compared to literature
(Provencher, 2001). (b) Amplitude estimation of metabo-
lites relative to Cre compared to literature (Cudalbu et al.,
2006; Pfeuffer et al., 1999).
00.511.522.533.54
0
0.02
0.04
0.06
0.08
0.1
0.12
0.14
0.16
0.18
ppm
Amplitude [a.u.]
Signal
Fit
Residual
(a)
0
5
10
15
20
25
30
35
40
ppm
Quality factor (Q)
3.9
3.7
3.5
3.3
3.1
2.9
2.7
2.5
2.3
2.1
1.9
1.7
1.5
1.3
1.1
0.9
0.7
0.5
0.3
0.1
Damp 0.01 − 12 metabolites
Damp 0.01 − 4 metabolites
(b)
Figure 5: Quantification analysis via the Quality factor (Q)
for the signal at 1.5 T using an incomplete basis set of
metabolites. (a) Fit using a basis set with 4 metabolites.
(b) Quality factor plot for the signal using a complete and
incomplete basis set (12 and 4 metabolites). The quality
factor computed for the signals was 2.1207 and 7.4472 re-
spectively.
(c) the 4 most relevant metabolites having the highest
concentration (Cre, GPC, Glu, NAA). Each basis set
is extended with the corresponding MM signal. The
lack of some metabolites in the basis set leads to a
00.511.522.533.54
−2
0
2
4
6
8
10
12
x 10
5
ppm
Amplitude [a.u.]
Signal
Fit
Residual
(a)
0
5
10
15
20
25
30
35
40
ppm
Quality factor (Q)
0.1
0.3
0.5
0.7
0.9
1.1
1.3
1.5
1.7
1.9
2.1
2.3
2.5
2.7
2.9
3.1
3.3
3.5
3.7
3.9
Damp 0.05 − 12 metabolites
Damp 0.05 − 4 metabolites
(b)
Figure 6: Quantification analysis via the Quality factor (Q)
for the signal at 9.4 T using an incomplete basis set of
metabolites. (a) Fit using a basis set with 4 metabolites.
(b) Quality factor plot for the signal using a complete and
incomplete basis set (16 and 4 metabolites). The quality
factor computed for the signals was 1.4033 and 5.1740 re-
spectively.
slight under- or overestimation of some other metabo-
lites and this is also reflected in the residual. Ampli-
tude estimates in Fig.4 are close to those presented
in literature, however, it is important to mention that
they are highly affected by individual conditions of
the tissues, small differences in the measurement pa-
rameters, size of the voxel measured and therefore di-
verse concentration ranges are found in similar stud-
ies, leading to a high variability of the amplitude es-
timates. In Fig.5 (a) and 6 (a) we observe that the
fits with an incomplete basis set are not good and the
corresponding Q
fit
of 7.4472 and 5.1740 also confirm
the imperfect quantification. In order to further eval-
uate the quantification, we selected a span or window
of 0.1 ppm to evaluate the quality factor in a moving
window. In Fig.5 (b) and 6 (b) we present the results
of this quality measure where the plotted curves rep-
resent the Q
fit
values obtained for all the frequency
intervals selected and the dashed line represents the
confidence bound calculated as three times the stan-
dard deviation (
R
2
N
> (3σ)
2
or equivalently Q
fit
> 9).
A missing metabolite is considered when the Q
fit
value is higher than the selected threshold.
5 CONCLUSIONS
Reliable metabolite estimation of in vivo MRS sig-
nals for determination of metabolite concentrations is
of paramount importance for obtaining additional in-
formation in the diagnostics of cancer and metabolic
BIOSIGNALS 2011 - International Conference on Bio-inspired Systems and Signal Processing
180

diseases. Therefore, quantification of MRS signals
was performed evaluating the influence of the damp-
ing factor constraint and the number of components
used in the metabolite basis set used for quantifica-
tion. We observed in particular, that the damping fac-
tor in the quantification method AQSES plays an im-
portant role in amplitude estimation. From the quan-
tification results, we examined the residual and ana-
lyzed the fit of the individual components which are
sensible to quantification constraints. The selection of
the metabolites for the basis set is important for quan-
tification, thus an incomplete basis set will provide fits
where one metabolite fits the region that corresponds
to its neighbor. The residual is used to determine the
goodness of the estimates. It is assumed that a good
estimate will lead to residuals resembling pure white
noise. A test of normality would also help to analyze
the residual and determine how close it is to white
noise.
ACKNOWLEDGEMENTS
Maria I. Osorio Garcia and Dr. Flemming U. Nielsen
are Marie Curie research fellows in the EU train-
ing network FAST (www.fast-mrs.eu). Dr. Diana M.
Sima is a postdoctoral fellow of the Fund for Sci-
entific Research-Flanders. Prof. Dr. Uwe Himmelre-
ich and Prof. Dr. ir Sabine Van Huffel are full pro-
fessors at the Katholieke Universiteit Leuven, Bel-
gium. Research supported by: Research Council
KUL: GOA Ambiorics, GOA MaNet, CoE EF/05/006
Optimization in Engineering (OPTEC), IDO 05/010
EEG-fMRI, IDO 08/013 Autism, IOF-KP06/11 Fun-
Copt, several PhD/postdoc & fellow grants; Flem-
ish Government: FWO: PhD/postdoc grants, projects:
FWO G.0302.07 (SVM), G.0341.07 (Data fusion),
G.0427.10N (Integrated EEG-fMRI) research com-
munities (ICCoS, ANMMM); IWT: TBM070713-
Accelero, TBM070706-IOTA3, TBM080658-MRI
(EEG-fMRI), PhD Grants; Belgian Federal Science
Policy Office: IUAP P6/04 (DYSCO, ‘Dynami-
cal systems, control and optimization’, 2007-2011);
ESA PRODEX No 90348 (sleep homeostasis), EU:
FAST (FP6-MC-RTN-035801), Neuromath (COST-
BM0601), KU Leuven center of Excellence ’MO-
SAIC’.
REFERENCES
Bottomley, P. (1984). Selective volume method for perform-
ing localized NMR spectroscopy. in U.S patent, (4 480
228).
Cudalbu, C., Cavassila, S., Ratiney, H., Grenier, D.,
Briguet, A., and Graveron-Demilly, D. (2006). Esti-
mation of metabolite concentrations of healthy mouse
brain by magnetic resonance spectroscopy at 7T.
Comptes Rendus Chimie, 9(3-4):534 – 538.
Grage, H. and Akke, M. (2003). A statistical analysis of
NMR spectrometer noise. Journal of Magnetic Reso-
nance, 162(1):176 – 188.
Gruetter, R. (1993). Automatic localized in vivo adjustment
of all first- and second- order shim coils. Magnetic
Resonance in Medicine, 29(6):804–811.
Klose, U. (1990). In vivo proton spectroscopy in presence
of eddy currents. Magnetic Resonance in Medicine,
14(1):26 – 30.
Laudadio, T., Mastronardi, N., Vanhamme, L., Van Hecke,
P., and Van Huffel, S. (2002). Improved Lanczos algo-
rithms for blackbox MRS data quantitation. Jorunal
of Magnetic Resonance, 157(2):292 – 297.
Pfeuffer, J., Tkc, I., Provencher, S. W., and Gruetter, R.
(1999). Toward an in vivo neurochemical profile:
Quantification of 18 metabolites in short-echo-time
1
H NMR spectra of the rat brain. Journal of Magnetic
Resonance, 141(1):104 – 120.
Pijnappel, W. W. F., van den Boogaart, A., de Beer, R., and
van Ormondt, D. (1992). SVD-based quantification
of magnetic resonance signals. Journal of Magnetic
Resonance, 97(1):122 – 134.
Poullet, J.-B., Sima, D., Simonetti, A., De Neuter, B.,
Vanhamme, L., Lemmerling, P., and Van Huffel, S.
(2007). An automated quantitation of short echo time
MRS spectra in an open source software environment:
AQSES. NMR in Biomedicine, 20(5):493 – 504.
Provencher, S. (2001). Automatic quantitation of local-
ized in vivo
1
H spectra with LCModel. NMR in
Biomedicine, 14(4):260–264.
Ratiney, H., Coenradie, Y., Cavassila, S., van Ormondt,
D., and Graveron-Demilly, D. (2004). Time-domain
quantitation of
1
H short echo-time signals: back-
ground accommodation. MAGMA, 16(6):284 – 296.
Slotboom, J., Nirkko, A., Brekenfeld, C., and van Ordmont,
D. (2009). Reliability testing of in vivo magnetic reso-
nance spectroscopy (MRS) signals and signal artifact
reduction by order statistics filtering. Measurement
Science and Technology, 20(104030):14pp.
Stefan, D., Di Cesare, F., Andrasescu, A., Popa, E.,
Lazariev, A., Vescovo, E., Strbak, O., Williams, S.,
Starcuk, Z., Cabanas, M., van Ormondt, D., and
Graveron-Demilly., D. (2009). Quantitation of mag-
netic resonance spectroscopy signals: the jMRUI soft-
ware package. Measurement Science and Technology,
20(10):104035(9pp).
Tk´aˇc, I., Starˇcuk, Z., Choi, I.-Y., and Gruetter, R. (1999). In
vivo
1
H NMR spectroscopy of rat brain at 1ms echo
time. Magnetic Resonance in Medicine, 41(4):649–
656.
DAMPING FACTOR CONSTRAINTS AND METABOLITE PROFILE SELECTION INFLUENCE MAGNETIC
RESONANCE SPECTROSCOPY DATA QUANTIFICATION
181
