
OPN-Ont: Object Petri Nets Ontology Tool
Lynda Dib and Fouad Bousetouane
LASE- LAboratoire des Systèmes Embarqués,
Computer Science Department, Badji Mokhtar University, Annaba, Algeria
Keywords: OPN-Ont System, Ontology, Objects Petri Nets, Requests, Concepts, Relations.
Abstract: Ontologies are being used nowadays in many areas, including software engineering, business, and biology,
to evaluate their suitability for representing and simulating domain processes. To assist users in developing
and maintaining ontologies a number of tools have been developed. The representation of knowledge bases
and conceptual domain models, hierarchical process, the structural components that participate in the
process and the roles that they play in a complex domain, is therefore a major challenge for computer
scientists for this complex domain. Without aiming at exhaustiveness, our study combining ontology and
Petri Nets (PNs) tries to identify some promising tracks in this area, which seems a rather interesting
alternative in the optics of the expressive power of the deductive representations. The context of our work
consists to develop a graphical knowledge model for complex domain. This paper presents the OPN-Ont
(Object Petri Nets Ontology) model. In this system ontology is represented in the PNs format, which allows
verification of formal properties and qualitative and quantitative simulation. It leads to represent and exploit
the different ontological components: concepts, relations and requests. The illustration of our model is made
in biological domain where process supports methods for qualitative and quantitative reasoning.
1 INTRODUCTION
In computer science, ontologies are a technique or
technology used to represent and share knowledge
about a domain by modelling the concepts in that
domain and the relationships between those concepts
(Gruber, 1991). These relationships describe the
properties of those concepts; in essence, what it is to
be one of those concepts in the domain being
modelled. Ontology represents a conceptualization
of reality or simply reality. Ontology often resorts to
various tools of formalization and representation,
which taken independently do not lead to the
anticipated results. Today, the number of tools for
developing ontologies has been increased and
diversified. Increasingly, the construction of
ontologies is an area of ongoing research. Today,
there are a number of models and tools for
developing ontologies. We assessed diverse models
that were developed in the fields of software
engineering, business, and biology, to evaluate their
suitability for representing and simulating domain
processes. Based on this assessment, we propose an
OPN-Ont model that should be mathematically
based to allow verification of properties that are
desirable in biological system, and simulation of
system behaviour. So, we have combined the best
aspects of two models ‘PN and ontology model’ and
we have developed OPN-Ont (Objects Petri Nets
Ontology). This last has an interactive graphical
interface based on Object PN. The illustration of our
model is made in biologic process where PNs can
represent nesting and ordering of biologic processes,
the structural components that participate in the
process and the roles that they play. OPN-Ont not
only represents hierarchical process knowledge in
biology (which is a major challenge for
bioinformatics) and structure components but it
composed queries to discover relationships among
processes and structural components. We used PNs
analysis to answer queries about the dynamic aspects
of the model. OPN-Ont is tested by representing
OntoCell (Dib, 2005), and composed queries to
discover relations among processes and structural
components. We used reachability analysis to
answer queries about the dynamic aspects of the
model.
2 RELATED WORK
There are many formalisms and tools to edit, browse
158
Dib L. and Bousetouane F..
OPN-Ont: Object Petri Nets Ontology Tool.
DOI: 10.5220/0004085301580163
In Proceedings of the 2nd International Conference on Simulation and Modeling Methodologies, Technologies and Applications (SIMULTECH-2012),
pages 158-163
ISBN: 978-989-8565-20-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

and map ontologies and a few comparative studies
of ontology tools have been performed. These tools
assist users in developing and maintaining
ontologies as: Workflow used by Peleg to develop
ontology fo biological processes (Peleg, 2002).
Other tools are developed to creat, edit and brows
ontologies such as: Protégé-2000; Ontolingua that
shows the concepts in a two dimensional tree
visualisation (Rice, 1996); Chimaera (Chimaera
Software Description URL, 2004) which is a web-
based ontology system and built on top of the
Ontolingua Distributed Collaborative Ontology
Environment; OilEd which uses the FaCT system
(The FaCT System URL, 2004) a description logic
system for checking the consequences of the
statements in the ontology and which has various
types of tabs where each tab shows information on
the current ontology component (Habbouche, 2002).
Evaluation: A number of other ontology tools
have been developed and used in bioinformatics.
However, only few evaluations of ontology tools
using bio-ontologies have been performed. In
(Lambrix, 2003), Protégé, Chimaera, and OilEd
were evaluated with respect to criteria such as
functionality, data model learning and user interface.
So, they were evaluated as ontology development
tools using GO ontologies as test ontologies. In
(Lambrix, 2004) an extension of this evaluation is
found, where Protégé-2000 with Chimaera were
evaluated against as ontology merging tools. In
(Dragan, 2006) the specific graphical user interface
provides graphical tools for all PN concepts and in
addition, the PN ontology is represented in RDFS,
and concrete PN models are represented in RDF.
However this solution covers only Time PNs, and no
other kinds of PNs. It neither defines PN structuring
mechanisms, nor provides precise constraints.
Finally, it does not enable using other ontology
languages for representing the PN ontology. From
this evaluation, no system is preferred but each
system has its own strengths and weaknesses. Based
on this assessment and on the solution proposed by
Perleg in (Pelegl, 2002), we combined ontology, PN
and a biological concept model and developed an
interactive graphical knowledge model, OPN-Ont,
tested in biological processes that supports methods
for qualitative and quantitative reasoning.
3 OPN-Ont
OPN-Ont model allows ontologies to be created and
explored. It is a computer application for data
organization and analysis. The OPN-Ont tool can
represent nesting and ordering of processes, the
structural components that participate in the
processes, and the roles that they play. It has an
interactive graphical interface based on high-level
PNs an extention of a PNs formalism. So, it maps to
PNs which is a graph-theoretical model that allow
verification of formal properties and qualitative
simulation. OPN-Ont tries to interpret all the
changes and states of the ontology being built or
operating. The user can interact with the system
using menus and graphical representation of
concepts and their relationships (in form of objects
PNs). Ontology in OPN-Ont system is operating by
updates and requests-answers trough queries. The
ontology is primarily an evolutes tool and the
updates must be performed periodically to adapt it to
its ontological function.
4 OPERATING SYSTEM
OPN-Ont
OPN-Ont provides both a net-based and a node-
based view of an ontology, where the latter displays
the selected concept and its entire environment
(definition, parents, children, other concepts linked a
domain relationship). So it is not limited to only
hierarchical link, is-a or part-of, however, the user
can hide links if they choose to. From the main
functions menu, the user can choose to:
Open/Creat/Save/Queries a ontology or Exit the
system. Once Open or New is chosen, the user could
introduce all the information (concepts and relations
that connect thems) collected and required for the
construction of the new ontology or the enrichment
of an existing one. At this level (Open/New), the
updates could be through a menu (Figure 1).
A-Consult: The system allows user to browse or
explore the ontology moving from one concept to
another. The marked place will be displayed with its
name and its entire environment : generic/specific
concepts and concepts that are linked by a domaine
relationship (Figure2).
The marked (current) concept can has equivalent
terms (called not-concepts or synonym concepts)
viewed in a sorted alphabetical list. As the not-
concepts are only linked to the current-concept and
can not referred to other concepts therefore their
representation in the PNs form is not essential and a
list of their names is more than sufficient. Only at
this level changes in the equivalent concepts can be
deleted, renamed or canceled.
b-Create: Through the creation menu the user
must specify if the relationship is equivalent,
specific, generic, or a domaine link. In the case of:
OPN-Ont: Object Petri Nets Ontology Tool
159
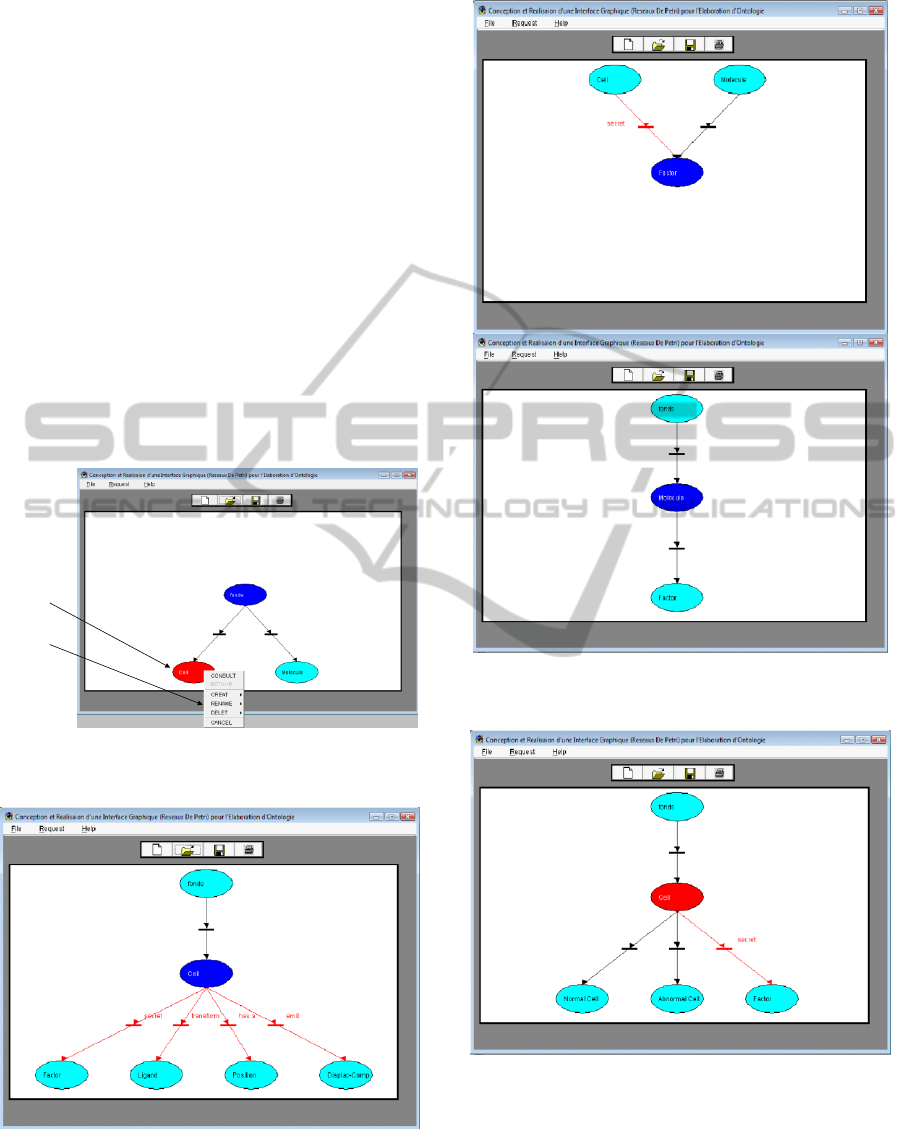
(a) generic and associative to a domaine links, the
name of the concept that the relation will be
established, should be included in the ontological
base, that means it must have at least one or more
generic links with one or more concepts (Figure 3,
Figure 4). (b) the specific link, the name of the
concept which will be linking may exist or not in the
ontology. If this name does not exist, a control
ensuring its no duplication is activated. At this stage,
the new concept can be added to the ontology
(Figure 5). (c) if the introduced name is a not-
concepts in the ontology, the system return
automatically to the concept which it is equivalent.
(d) the creation of a not-concept, the system control
ensuring the non membership of this term to the list
of concepts or to the list of not-concepts. To add a
new link the system always ensures it has not been
duplicated. Thus, there will be a base of information
that respects the principles of ontology
establishment.
Concept
(Marked place)
Concept Menu
Figure 1: Concept menu: Menu of possible operations
associated to a marked place.
Figure 2: Consultation of the current-concept.
Figure 3: Creation of a generic relationship between
‘Molecule’ and ‘Factor’ concepts in OntoCell ontology.
Figure 4: Creation of a domain relationship: Cell ‘secret’
Factor.
C-Renaming Concept or Relationship
Through the ‘Rename Menu’ the user can rename a
concept or a not-concept but it is necessary to avoid
redundancy. He can also renaming a domain
relationship among concepts.
SIMULTECH 2012 - 2nd International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
160
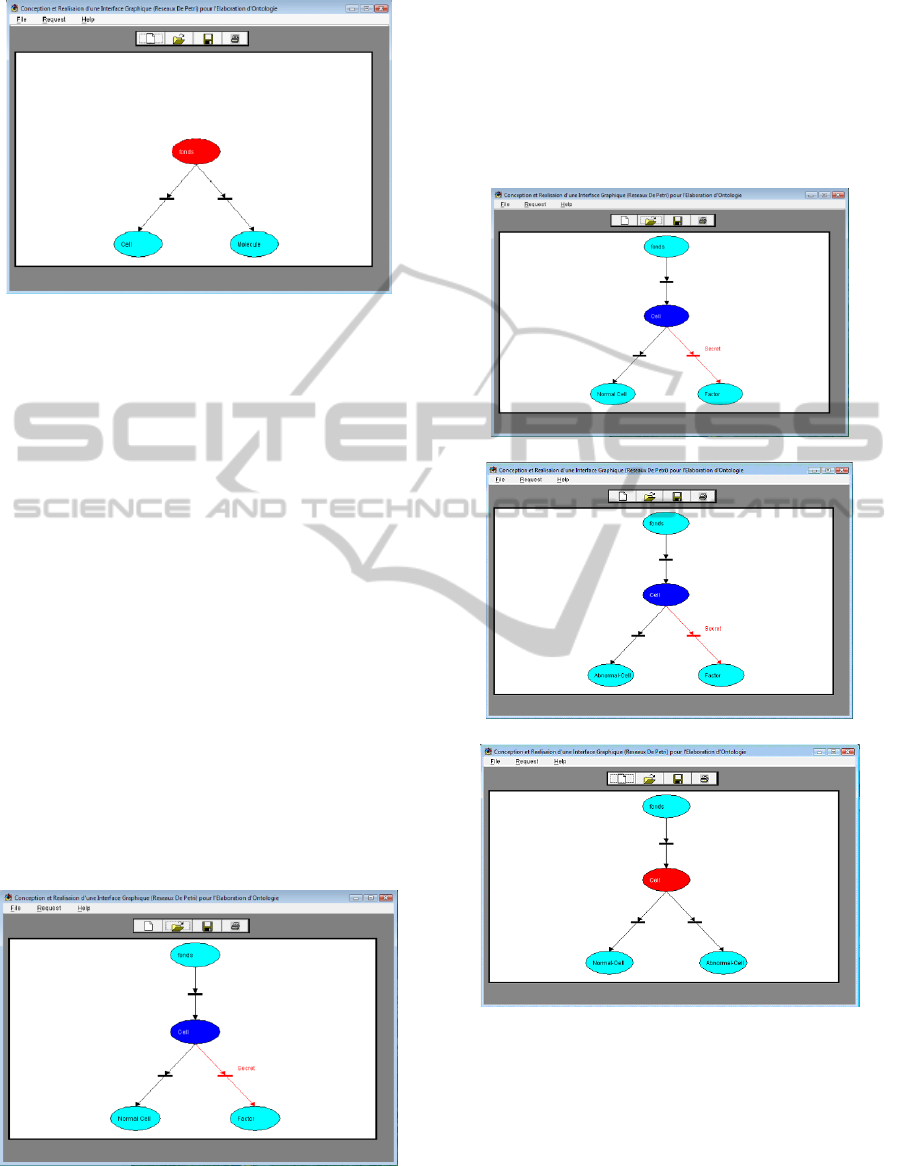
Figure 5: Creation of a specific relationship: Creation of
‘Cell & Molecule’ concepts in OntoCell ontology.
D-Delete: Through the ‘Delete Menu’ the user can
delete a concept previously introduced (error during
the introduction of the concept, ...). After
confirmation, the system removes all links that relate
the concept on cause to other ones Figure 6.
If further, one of specific concepts (concept2) to
the removed concept (concept1) was not attached to
any other concept, the elimination of concept1 will
cause automatically the deletion of concept2 if the
user wants this, otherwise he will relate concept2 to
another existing concept in the ontology to avoid
any loss of information. This principle will be
applied to specific concept of concept2 and so on
until to the last specific one. We note that deleting
only a relation (with a specific (Figure 7a), generic
(Figure 7b), or concept linked by a domain
relationship (Figure 7c), between concept2 and
concept1 and not a related concept, is possible by
selecting the relation in cause and choosing the
Delete option. The same verification process
described above will be followed if concept2 is not
connected to any other concept that concept1.
Figure 6: The result after deletion of a concept.
3 - Requests: Once the ontology is built, queries
about its contents may be made. Once the user
describes this request by entering the operators (OR,
AND, EXCEPT) and their inputs, the system uses
the appropriate processing with the possibility of
combining them. The user structures his query as a
PN form starting with the most general operator of
the query to the more specific one.
a: Delete a specific link.
b: Delete a generic link.
c: Delete a domain link.
Figure 7: Examples of relationship delation.
5 RESULTS AND DISCUSSION
Results: A biological OPN-Ont Example
The example represent OntoCell ontology (Dib,
2005). Due to lack of space we only show some
concepts of OntoCell diagrams. In the first part of
OPN-Ont: Object Petri Nets Ontology Tool
161
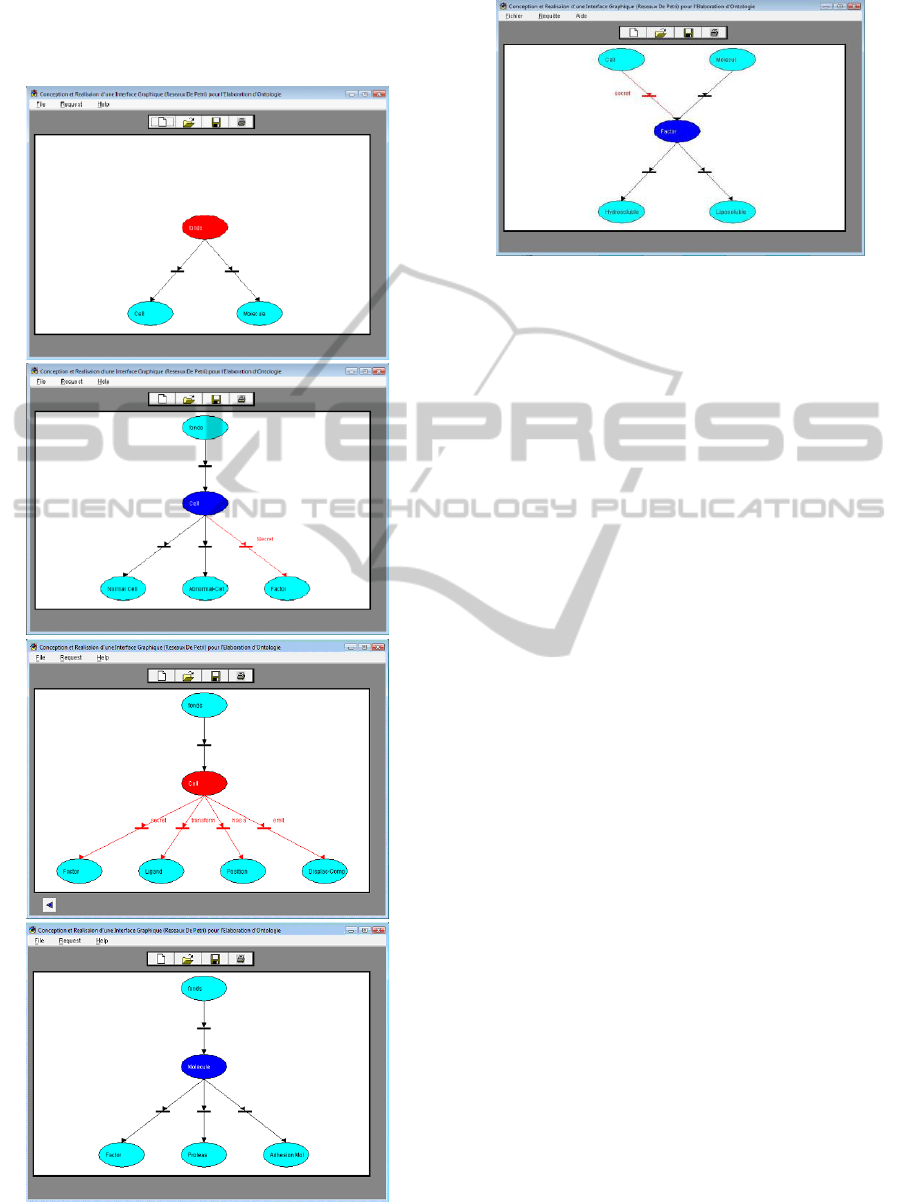
this example we represent OntoCell Concepts.
Initially, we represent two concepts (Cell and
Molecule) of the first level of OntoCell (Figure 8 a).
(a)
(b)
(c)
(d)
Figure 8: Some concepts in OntoCell ontology.
(e)
Figure 8: Some concepts in OntoCell ontology(cont.).
After, we represent the second level, where we
represent for example the specific (black transition)
and associative to a domain (red transtion) links of
Cell concept (Figure 8 b,c) and specific Molecule
concept (Figure 8 d). At last, in the third level, we
represent for example the specific concepts of the
concept Factor (Figure 8 e). Factor is linked to
Molecule by a specific relation and to Cell by the
role ‘Secret’.
In the second part we identify queries. The
results of the three first queries are given by Figure
9: Case a, shows the result generated by the query
‘Cell AND Molecule’. Case b, shows the result
generated by the query ‘Cell OR Molecule’. Case c,
shows the result generated by the query ‘Cell
EXCEPT Molecule’.
Discussion
OPN-Ont allows Ontologies to be created and
explored. Places represent concepts, and transitions
represent relationships (hierarchical links or
activities). There are many benefits to use PNs: they
have a firm mathematical foundation and they
explicitly represent states, which allows for the
modeling of milestones and implicit choices.
Another benefit is that Hierarchical PNs can control
the complexity of the representation of biological
systems. And last, Colored PNs can define states and
transitions and dynamical behaviours of the systems
are indicated by distributions of tokens changed
progressively along individual fulfilments of
conditions at places and succeeding firing the events
at transitions.
SIMULTECH 2012 - 2nd International Conference on Simulation and Modeling Methodologies, Technologies and
Applications
162
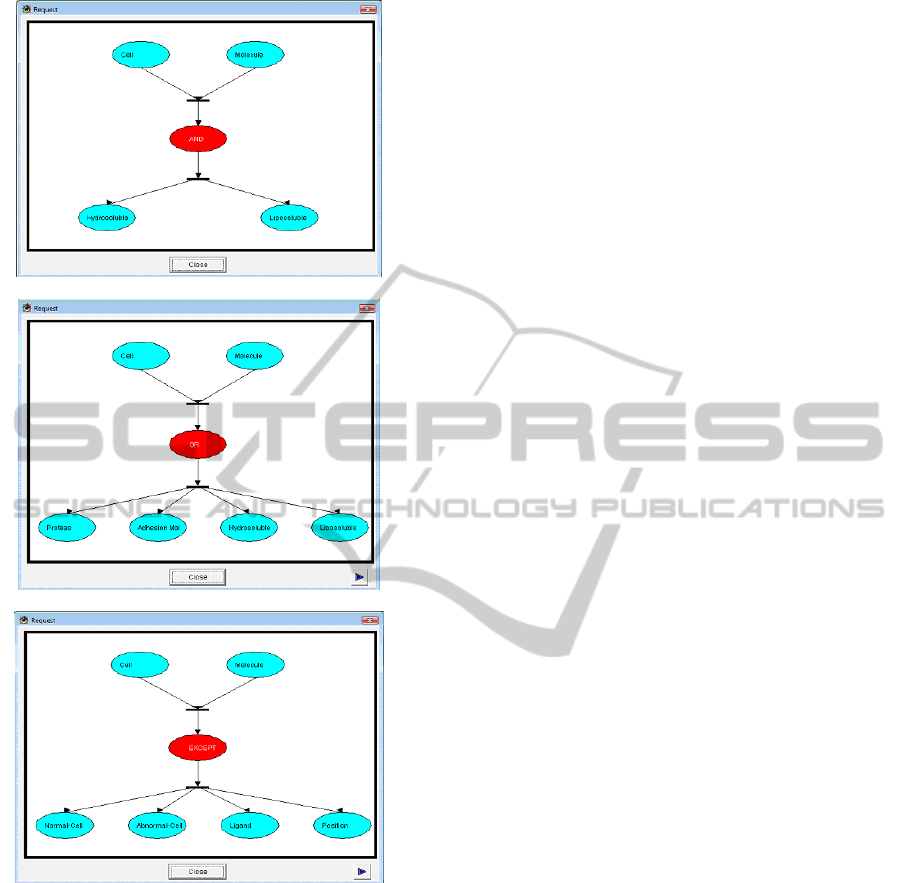
Response of query: 'Cell AND Molecule'.
Response of query: 'Cell OR Molecule'.
Response of query: ’Cell EXCEPT Molecule'.
Figure 9: OntoCell in OPN-Ont: functional role, biological
reactions, biological process.
6 CONCLUSIONS
The aim of OPN-Ont tool is to allow experts, each in
its area, to construct and operate their ontologies.
The graphics offers the user ease of handling and
understanding the behaviour of the system by the
various commands he sends. The search for
information thus becomes easy, either by direct
access of the system to the information requested
through requests or by exploring the ontology while
navigating the network that represents it. PNs were
chosen to model ontology; indeed, knowledge is
clearly represented and easily identified. Also, the
dynamic aspect will be present in the activity of
conceptual knowledge and in requests where the
evolution of marks in places facilitate for the system
the search of the requested information, especially in
the search for intersection among two distant
concepts in the network but that have temporarily
dynamic link (with operators) which disappears after
reply to the request.
REFERENCES
Chimaera Software Description URL, 2004:
www.ksl.stanford.edu/software/chimaera (Acc.25-07).
Dib L., 2005: OntoCell: An ontology of Cellular Biology,
Journal of Computer Science 1 (3): 445- 449.
Gruber, T. R., 1991: The role of common ontology in
achieving sharable, reusable knowledge bases, in
Proceedings of KR'1991: Principles of knowledge
representation and reasoning, p. 601-602.
Habbouche, M. and Pérez, M. 2002: Evaluation of
ontology tools for bioinformatics, LiTH-IDA-Ing-Ex-
02/10, Department of computer and information
science, Linköpings universitet. In Swedish.
Lambrix, P., Edberg, A., 2003: Evaluation of ontology
merging tools in bioinformatics, Pacific Symposium on
Biocomputing, 8:589-600, Kauai, Hawaii, USA,
Lambrix, P., Habbouche, M., Pérez, M., 2004: Evaluation
of ontology development tools for bioinformatics,
Bioinformatics, 19(12):1564-1571.
Rice, J., Farquhar, A., Piernot, P.and Gruber, T. 1996:
Using the Web Instead of a Window System. In CHI,
pp.103–110. http://www-ksl-svc.stanford.edu: 5915/
doc/papers/ksl-95-69/index.html.
The FaCT System URL: 2004 www.cs.man.ac.uk/~
horrocks/FaCT. (Acc. 21-07).
Dragan G., Vladan D. 2006: Petri net ontology, in
Knowledge-Based Systems Journal. 19 220–234
M. Peleg, I. Yeh, R. Altman, 2002: Modeling biological
processes using Workflow and Petri net models, in
Bioinformatics Journal 18 (6) 825–837.
(a)
(c)
(b)
OPN-Ont: Object Petri Nets Ontology Tool
163
