
A Grid based Medical Image Retrieval System using Alchemi
F. Maghraby
1
, H. M. Faheem
2
, M.
Roushdy
2
and M. Amoon
3
1
Higher Institute of Computer and Information Technology, ELShorouk Academy, ELShorouk City, Egypt
2
Faculty of Computer and Information Sciences, Ain Shams University, Abbassia, Cairo, Egypt
3
Faculty of Electronic Engineering, Menoufia University, Shebeen El-Kom, Menufia, Egypt
Keywords: Alchemi, Database Partitioning, Dicom, Grid Computing, Semantic Features.
Abstract: This paper proposes an approach to perform retrieval process on medical image databases by extracting
semantic information from the dataset values of the DICOM (Digital Imaging and Communications in
Medicine) format which produces a set of images relevant to the given query. Image retrieval in general has
the goal to allow for the retrieval of similar images over very heterogeneous image collections to help the
diagnostic process. With modern radiology, departments produce tens of thousands of images per day. It is
apparent that infrastructures are required to treat this large amount of data. Grid technologies are among
those approaches deployed to make computing power available to large-scale research projects. Often, the
goal is to have a very large number of resources in various locations that can be shared for performing
computationally intensive tasks. Grid computing has the potential to help computer science researchers in
medical institutions to better use an existing infrastructure. It shows that particularly computationally–
intensive tasks such as the extraction of features from large image databases can be performed much faster.
Alchemi framework has been deployed in this paper to provide grid-based environment .Speeding up the
retrieval process was one of the major achievements of this work.
1 INTRODUCTION
Computer grids are promising architectures with a
strong potential for sharing resources. They are
generally valued for the large computing power and
data storage space they provide. Beyond this
interest, grid technologies allow scientists federated
in Virtual Organizations (VOs) to easily share
datasets and algorithms across boundaries of their
organizations. All these grid characteristics make
them particularly interesting for the medical
community who deals with large and fragmented
amounts of medical images. As a consequence,
various medical images simulation, storage, and
processing applications have recently been
developed on grids (
Montagnat et al., 2004b). The
problem of large scale image indexing and retrieval
remains relevant for many of them.
The proposed system uses Alchemi which is an
open source software framework that can be
deployed to aggregate the computing power of
networked machines into a virtual supercomputer
(desktop grid) and to develop applications to run on
the grid. The proposed system uses the DICOM
information for performing the retrieval on medical
images. The retrieval is performed by extracting
semantic features from the dataset values of the
DICOM format. The extracted information can be
used to perform the retrieval which produces a set of
images relevant to the given query.
The rest of this paper is organized as follows:
section 2 provides a brief introducing to general grid
computing principles. Section3 explains database
partitioning on grid. Section 4 discusses content
based image retrieval. Section 5 presents our
proposed system and its modules. Section 6
discusses the experimental results. Section 7
provides some concluding remarks.
2 GRID ENVIRONMENT
Computer grids consist of a network of computers
providing distributed computing and storage
resources to their users through a grid middleware.
The middleware is the software layer implementing
basic services to access a grid infrastructure and
hiding the system complexity to the user (Camarasu
224
Maghraby F., M. Faheem H., Roushdy M. and Amoon M..
A Grid based Medical Image Retrieval System using Alchemi.
DOI: 10.5220/0004448202240230
In Proceedings of the 15th International Conference on Enterprise Information Systems (ICEIS-2013), pages 224-230
ISBN: 978-989-8565-59-4
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
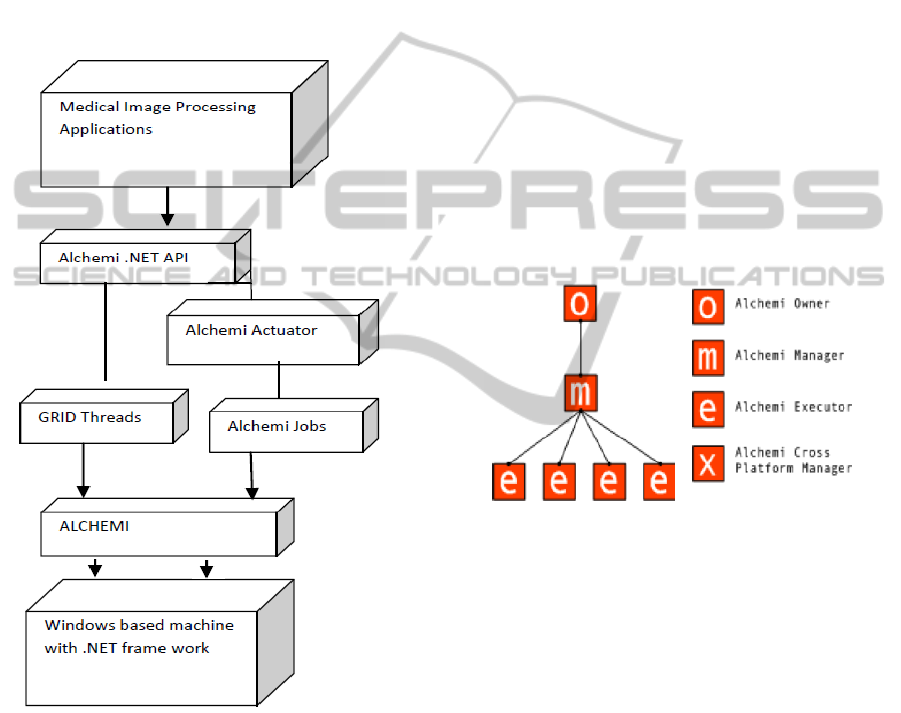
et al., 2008). Alchemi is one of the Software
frameworks to enable grid computing. It has been
primarily written for UNIX operating systems.
However, Microsoft .Net framework provides a
platform to implement windows based grid
computing environment with Alchemi as shown in
Figure1. In particular, it also provides remote
execution, security, multithreading,, asynchronous
programming, disconnected data access, and
managed execution. This makes it an ideal platform
for grid computing middleware (Dhivya and Ruba,
2012).
Figure 1: Layered Architecture of distributed Windows.
2.1 Architecture
Alchemi (Luther et al., 2005) follows the master-
worker parallel programming paradigm in which a
central component dispatches independent units of
parallel execution to workers and manages them.
This smallest unit of parallel execution is a grid
thread. A grid application is defined simply as an
application that is to be executed on a grid and that
consists of a number of grid threads. Grid
applications and grid threads are exposed to the grid
application developer via the object- oriented
Alchemi .NET API. Alchemi tool describes the four
components. They are:
Manager
Executor
Cross-platform manager
Owner
These components allow Alchemi to be utilized
to create different grid configurations desktop
cluster grid, multi cluster grid, and cross-platform
grid (global grid). (Dhivya and Ruba, 2012)
2.1.1 Cluster Desktop Grid
The cluster desktop grid (shown in Figure 2),
consists of a single Manager and multiple Executors.
One or more Owners can execute their applications
on the cluster by connecting to the Manager. Such
environment is appropriate for the deployment on
Local Area Networks as well as the Internet.
(Dhivya and Ruba, 2012).
Figure 2: Cluster (Desktop Grid) Deployment.
2.1.2 Multi Cluster Deployment
A multi-cluster environment (shown in Figure 3) is
created by connecting Managers hierarchically .As
in a single-cluster environment, any number of
Executors and Owners can connect to a Manager at
any level in the hierarchy. The key to accomplishing
multi-clustering in Alchemi's architecture is the fact
that a Manager at a given “intermediate” level is
treated by the higher level-Manager as an Executor.
Such an environment is more appropriate for the
deployment over the Internet.
2.1.3 Cross-Platform Manager
A grid middleware component such as a broker can
use the Cross-Platform Manager (Shown in Figure
4) web service to execute cross-platform
applications (jobs within tasks) on an Alchemi node
(cluster or multi-cluster) as well as resources grid-
AGridbasedMedicalImageRetrievalSystemusingAlchemi
225
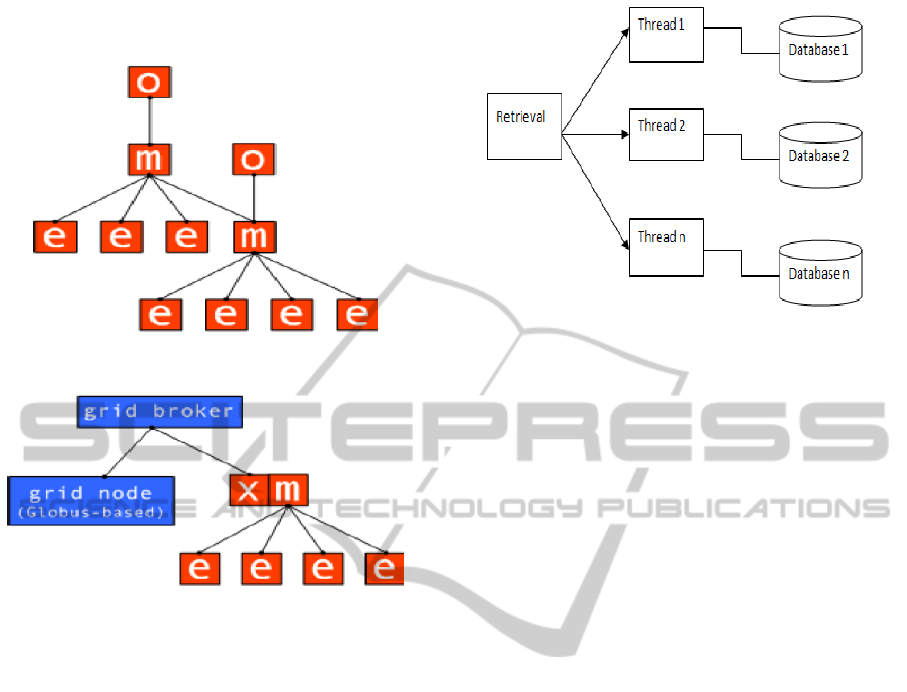
enabled using other technologies such as Globus.
(Dhivya and Ruba, 2012).
Figure 3: Multi Cluster Deployment.
Figure 4: Cross-Platform Manager.
3 DATABASE PARTITIONING
In this paper we study the impact of executing a
medical image database query application on the
grid. For lowering the total computation time, the
image database is partitioned in equal subsets to be
processed on different grid nodes. A theoretical
model of the application computation cost and
estimates of the grid execution overhead are used to
efficiently partition the database. Smart partitioning
of the database can lead to significant improvements
in terms of total computation time (Montagnat et al.,
2004).
If the database is partitioned in bags of images to
be analyzed, each bag can be analyzed by a single
computing job. If one bag is representing one image
so that all images could be processed in parallel,
then Alchemi threads could work in parallel. Hence,
the execution time would be the maximum of the
execution times of each image processing. See
Figure 5.
Figure 5: Database Partitioning Process.
4 CONTENT BASED IMAGE
RETRIEVAL
A large number of medical images in digital format
are generated by hospitals and clinics every day.
Such images constitute an important source of
anatomical and functional information for diagnosis
of diseases, medical research, and education. It is
well known that medical image databases are the
key component in diagnosis and preventive
medicine. This increasing trend towards digitization
of medical images creates a need of technologies for
storage, organization, and retrieval of the medical
images. Content based image retrieval (CBIR) is the
digital image searching problem in large databases
that makes use of the contents of the images
themselves rather than relying on the textual
information. These techniques use the automatically
derived features (low level feature) such as color,
texture and shape as search criteria. Medical images
generated in hospitals contain semantic information
(high level feature). This information can be used to
retrieve the images.
4.1 Semantic Feature Extraction
The DICOM standard was created by the National
Electrical Manufacturers Association (NEMA) to aid
the distribution and viewing of medical images, such
as CT(Computed Tomography) scans, MR(Magnetic
Resonance), and US (Ultrasound). Imaging
equipment used in hospitals generates images which
are in DICOM format. It is a standard format used to
obtain, store and distribute medical images. DICOM
comprise standardized textual descriptions of study,
patient, body region examined and modality. A
ICEIS2013-15thInternationalConferenceonEnterpriseInformationSystems
226
single DICOM file contains both a header (which
stores information about the patient's name, the type
of scan, image dimensions, etc), as well as all of the
image data. This is different from the popular
Analyze format, which stores the image data in one
file (*.img) and the header data in another file
(*.hdr). The DICOM header size varies depending
on how much header information is stored. The
header describes the image dimensions and retains
other text information about the scan. DICOM files
are composed by one image and tags describing the
image. Tags are textual or numerical sequences of
<attribute, value> pairs. The textual information is
considered as the semantic information. For all the
DICOM files the image and the relevant tags are
extracted and are stored in the database. The image
is stored in jpeg file format. The extracted semantic
information is stored in the database which is used
during the retrieval process. (Selvarani and
Annadurai, 2007).
4.2 Content Feature Extraction
Content Based Retrieval system represents each
image as a feature vector and measures the similarity
between images as the distance between their
corresponding feature vectors. For medical images,
shape and texture are the two important low level
features which describe the content of the image.
The shape and texture features are extracted and
stored in the database as feature vectors. (Selvarani
and Annadurai, 2007).
5 PROPOSED SYSTEM
5.1 Semantic Feature Extraction
Module
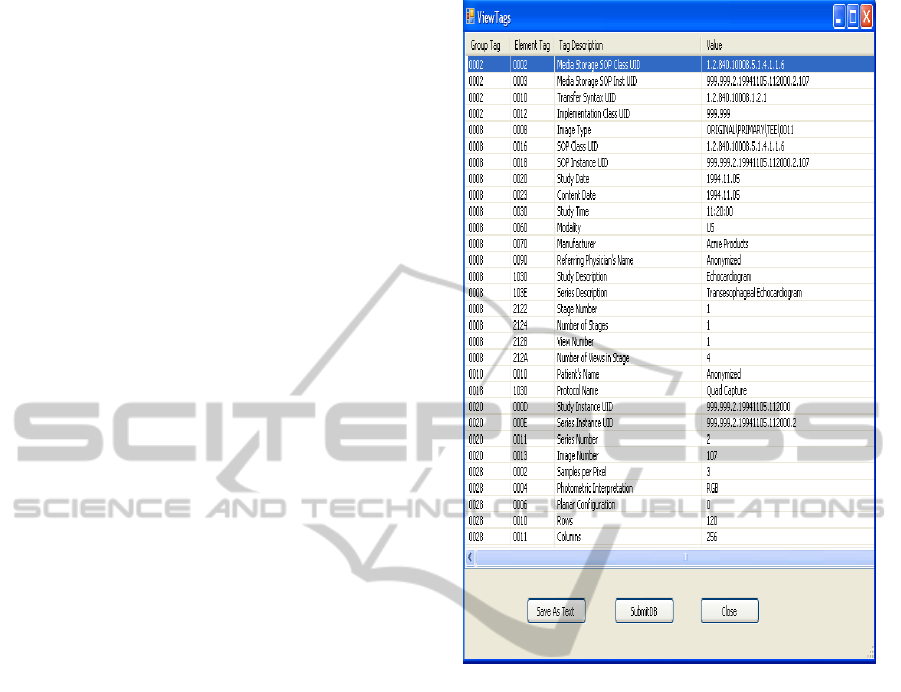
Semantic features module extracts DICOM tags
information from .dcm files format. Some extracted
DICOM information can be observed in Figure 6.
After extracting all needed features from images,
it will be stored in SQL database server to be used in
searching process, and then the database is
transported to computation nodes and partitioned to
equally sized subsets.
5.2 Database Modules
The database consists of 2 parts: DICOM images
and DICOM tags (Semantic Feature).
Figure 6: DICOM Tags.
5.2.1 DICOM Images
We used a database of 100,000 images from 6000
patients. The selected images were composed by CT
and MR images representing different anatomical
structure: Head, heart, Shoulders ..., etc. DICOM
images are stored as image data types, which are
data types that hold any type of binary data. We read
BLOBs (Binary Large Objects) as streams and
manipulate/display images according to the
information (e.g., Bit Depth) extracted from the
DICOM Info file.
5.2.2 DICOM Tags
For each DICOM image we extract all DICOM
semantic features and store it in SQL database and
make relation between image and its semantic
information for the ease of retrieval process.
5.3 DICOM Selection Module
In this module, the user interface guides user to
AGridbasedMedicalImageRetrievalSystemusingAlchemi
227
construct the query performing the selection by
querying DICOM tags. The system then selects the
most appropriate images related to these tags. The
selection is based on a simple SQL database query.
5.4 Output Module
In this module, the proposed system collects the
resulted images from all threads and then displays it
on the screen .Also all information related to the
resulted images are available to users who can save
it in a text or xml file format.
6 EXPERIMENTAL RESULTS
AND DISCUSSION
We construct Dictionary containing all DICOM tags
to allow users to choose tags related to their query
attributes. Assume that the user query is to retrieve
all the images of US modality. The user must
specify the query attributes (e.g., Acquisition Date,
Modality, Patient Name, Admitting Diagnosis
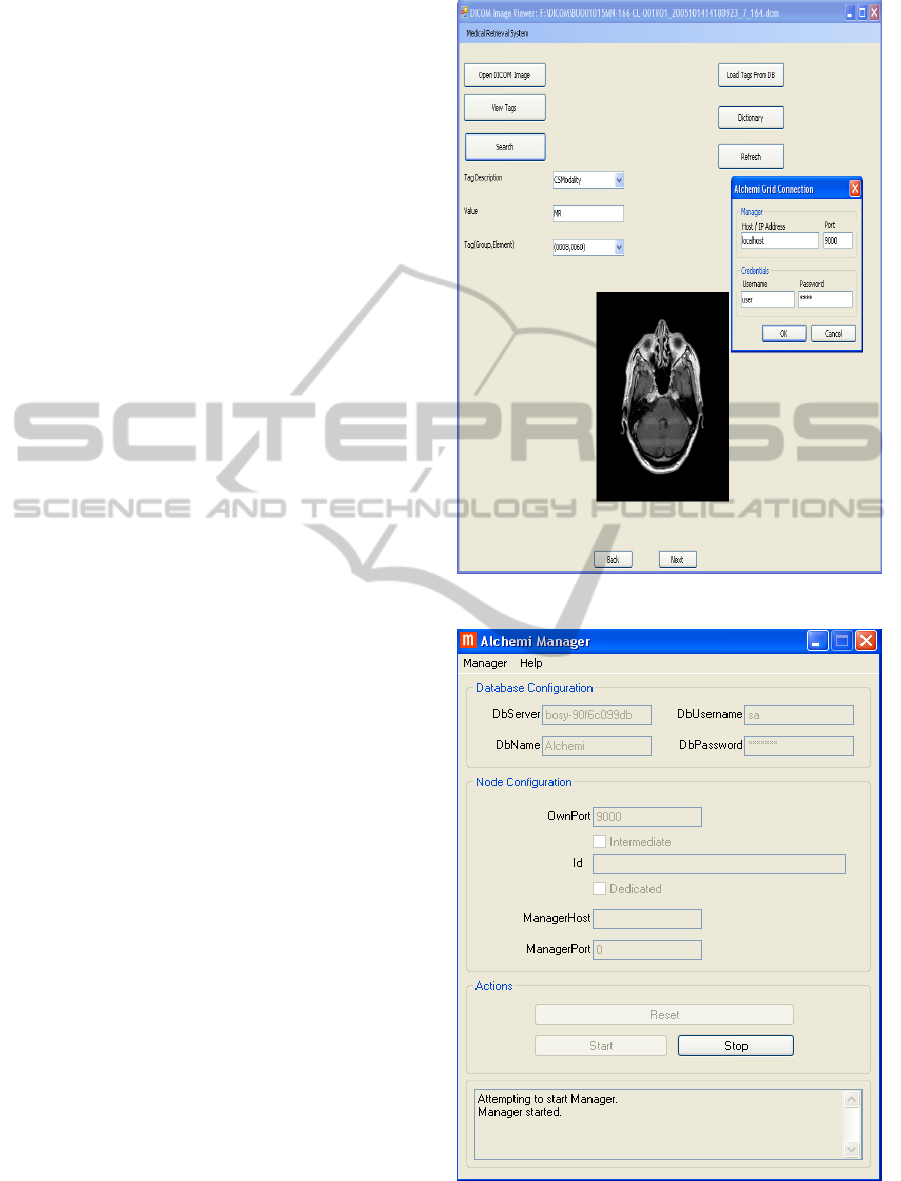
Description) using interface module. This is shown
in Figure 7.
When the user selects the search operation, the
system connects to grid middleware, and then
Alchemi imitates traditional multi threaded
programming. GThread is a grid thread and
GApplication is application thread. “Just in time
scheduler algorithm” is used for splitting the
comparison job into different threads. Assume that
T1, T2, T3….Tn are the threads generated by
Alchemi. Alchemi owner provides an interface with
the grid application between the application
developer and the grid. The owner submits the
completed threads to the Alchemi Manager. The
Alchemi Manager manages the execution of threads
responsible for the searching process in its own
dataset.
The executors register themselves with the
Manager which in turn keeps track of their
availability. Threads received from the Owner are
placed in a pool and scheduled to be executed on the
various available Executors. The Executor accepts
threads from the Manager and executes them.
Executor API is used to make an interface with the
Alchemi Manager.
Figure 8 shows the configuration of Alchemi
Manager. In the figure, setup connection enables or
starts the Alchemi manager. Figure 9 shows the
configuration of Alchemi Executor that contains the
host and the port number of connectivity, and
Figure 7: User Interface.
Figure 8: Manager Window.
ICEIS2013-15thInternationalConferenceonEnterpriseInformationSystems
228
Figure 9: Executor Window.
credentials required to login on to the system. Figure
10 shows Console Form .This form provides the
system statistics and real time graph of power
availability and usage.
We can see that partitioning the dataset on
different executor nodes while there is no task
dependency can lead to a significant improvement in
the retrieval time. Figure 11 provides a comparison
between sequential and parallel execution time. The
system was tested on set of 100,000 images .We had
provisioned 10 executor nodes.
Processing sequentially can be an inefficient way
to handle large datasets. The gap in runtime greatly
increases as the dataset grows larger in size .These
results demonstrate that the grid computing can
dramatically reduce the time required to retrieve
medical images.
7 CONCLUSIONS
In this paper we propose a grid based system for
retrieving medical images. Grids are promising
architectures that can bring different solutions to
medical image storage and retrieval problems. The
proposed system performs content based medical
Figure 10: Console Form.
Figure 11: Comparison between Sequential and Parallel
Execution Time.
image retrieval by extracting semantic information
from the dataset values of the DICOM format which
produces a set of images relevant to the given query.
To speedup retrieval process, we partitioned the
database into equally sized subsets. So, queries can
benefit from the grid computing parallelism and
AGridbasedMedicalImageRetrievalSystemusingAlchemi
229

execute different queries, each on a different subset
of images. We can see that Alchemi framework is
ideally qualified to be deployed in medical image
retrieval system .Of course, as the number of
executors increases, the overall system performance
increases. The system is considered a step towards a
complete grid-based implementation for a complete
medical retrieval system.
REFERENCES
Camarasu, S. , Benoit-Cattin, H. , Montagnat, J., and
Racoceanu, D., 2008,Content-Based Medical Image
Indexing and Retrieval on Grids, First International
Symposium on ICT for Health, Ateneo de Manila
University, Manila, Philippines, Philippine J Info
Tech.
Dhivya, M., Ruba, K., 2012, Building Grid based
Application for the Management of Medical Image
Data using Alchemi, International Conference on
Recent Trends in Computational Methods,
Communication and Controls (ICON3C 2012)
Proceedings published in International Journal of
Computer Applications® (IJCA).
Luther, A., Buyya, R., Ranjan, R., and Venugopal, S.,
2005, Alchemi: A .NET-based Grid Computing
Framework and its Integration into Global Grids,
GRIDS Lab, Australia.
Montagnat, J., Breton,V. and Magnin, I. , 2004,Medical
image databases content-based queries partitioning on
a grid, HealthGrid'04, Clermont- Ferrand.
Montagnat, J. and al, e., 2004, Medical image content-
based queries using the Grid, Proceedings of the first
European HealthGrid conference.
Selvarani, A. G., and Annadurai, S., 2007,Medical Image
Retrieval by Combining Low Level Features and
DICOM Features, Conference on Computational
Intelligence and Multimedia Applications,IEEE.
Tweed, T. and Brunie, S., 2003, Medical Image Database
on the Grid: Strategies for Data Distribution,
HealthGrid'03, Lyon.
ICEIS2013-15thInternationalConferenceonEnterpriseInformationSystems
230
