
Deformable Muscle Models for Motion Simulation
Tomáš Janák and Josef Kohout
Department of Computer Science and Engineering, University of West Bohemia,
Univerzitní 8, 30100, Plzeň, Czech Republic
NTIS – New Technologies for the Information Society, University of West Bohemia,
Univerzitní 22, 30100, Plzeň, Czech Republic
Keywords: Musculoskeletal Model, Medical Simulation, Soft-Body, Deformable Objects, Collision Detection, Mass-
Spring System.
Abstract: This paper presents a methodology for interactive muscle simulation. The fibres of individual muscles are
represented by particles connected by springs, thus creating a deformable model of the muscle. In order to
be able to describe human musculoskeletal system, contact between pairs of muscles as well as muscles and
bones must be accounted for. Therefore, collision detection and response mechanism which allows both
types of contact (soft body vs. rigid body and soft vs. soft body) is presented. The solution is a part of a pro-
ject dedicated to improvement of the effectiveness of osteoporosis prediction and treatment.
1 INTRODUCTION
The musculoskeletal system of modern human is
often a subject to various medical conditions, from
minor aches to serious diseases such as osteoporosis.
To improve the efficiency of treatment, virtual mod-
els can be used to simulate motion of the patient
during some situations (walking, jumping etc.) in
order to better understand how such workload af-
fects muscles and bones in these situations. The
creation of the whole musculoskeletal model that
could be tuned to a specific patient is a complicated
procedure with many steps that are outside the scope
of this paper. The focus here is solely on how to
model muscles during the simulated motion, assum-
ing their initial geometry is given.
In terms of computer simulation, a muscle is a
typical example of a soft-body object, i.e. its shape
is elastically changing in response to external forces.
There are two general approaches for simulating the
musculoskeletal system in motion. The first, simpler,
approach defines the animation by movement of the
bones and then deforms the muscles according to the
interaction with bones or potential obstacles. That
means that the muscles are deformed as a result of
the animation instead of being the initiators of the
animation. The second approach models individual
muscles as they actually work in real world, i.e. they
are the initiators of forces that move the bones. The
source of the animation then are the forces acting on
the bones produced by individual muscles. Although
the second approach is possible to use (Lee et al,
2009), it is obviously computationally much more
demanding and will not be considered here.
As the simulated patient moves through the vir-
tual scene, the muscles interact with the bones, pos-
sible obstacles in the scenes and also each other.
Therefore, apart from the obvious need to be able to
update the geometry in every step of the simulation,
the crucial part of the muscle simulation is efficient
collision detection, which identifies parts of the
muscle geometry that should be updated. There are
many different problems in computer graphics and
related fields which rely on collision detection.
However, in most cases the situation is slightly less
complicated and satisfied with rigid object vs. rigid
object, or one rigid vs. one soft-body object collision
detection. In the case of musculoskeletal system
simulation, all possible combinations of collision
situations happen at the same time. Furthermore,
each muscle is modelled separately. Therefore, the
neighbouring muscles are in fact in a state of contin-
uous collision, affecting each other in every single
step of the motion. That results in the need for a
robust collision handling methodology.
One could argue that all the adjacent muscles
could be treated as one large lump of tissue, but that
would limit the options the solution provides. First,
the operator will often be interested in analysis of
only a single (or several) muscles and lumping all
the muscles together would complicate that. Also,
301
Janák T. and Kohout J..
Deformable Muscle Models for Motion Simulation.
DOI: 10.5220/0004678903010311
In Proceedings of the 9th International Conference on Computer Graphics Theory and Applications (GRAPP-2014), pages 301-311
ISBN: 978-989-758-002-4
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

even non-adjacent muscles can touch each other
during various motions (e.g. thighs and calves touch-
ing while kneeling etc.) and therefore the collisions
would have to be checked for and treated neverthe-
less. Lastly, connecting individual muscles together
does not comply with the physical reality. Even
when attached to the same bone, muscles can slide
over each other and change their relative positions
by a significant margin. This would be difficult to
simulate if the muscles were as one.
The idea behind this work was to create a
framework that would enable medical operators to
quickly create an interactive simulation of motion,
i.e. ideally a real-time simulation, but at least a
frame every few seconds. Such simulation would
then be used for a coarse assessment of the situation
at hand and only after finding out the most critical
points during the motion, a more accurate (but also
time consuming) algorithm would be used for a
precise evaluation of those few points. Hence, even
though the simulation has to be realistic in order to
be useful, it does not aim to be perfect. The frame-
work was created as a part of the EU-funded
VPHOP project (www.vphop.eu), which is aimed on
developing technologies for better prediction of
bone fracture risks in order to be able to provide
more effective treatment of osteoporosis.
On the following pages, the paper will present
the created framework. There is no groundbreaking
new algorithm presented, rather the contribution is
in assessment of existing algorithms of soft-body
simulation and collision detection and “tweaking”
those for the purposes of the particular problem
described above.
This paper is structured as follows. After a brief
summary of the previous work done in related fields
in Section 2, the main part of the paper will follow
with detailed description and some implementation
details of the used muscle model and collision han-
dling mechanism (Section 3). Section 4 will con-
clude with experimental measurements of the
framework's performance.
2 STATE OF THE ART
2.1 Soft-Body Models
A soft-body represent a stiff, but deformable object.
The shape of the object changes according to exter-
nal forces, but at the same time the object resists
those forces and tries to maintain its original (“rest”)
shape. In general, soft-body models can be classified
as either heuristic or continuum mechanics, depend-
ing on whether the model behaves according to
actual elasticity principles or some heuristic that
aims only to produce a plausible, although not phys-
ically accurate, visualisation. A continuum mechan-
ics approach offers better fidelity and also more user
comfort because all the parameters of the model are
actual measurable physical properties. Heuristic
approaches usually require a lot of experimenting to
find suitable parameters, but they are significantly
faster during the run-time of the simulation. The
most common method for the continuum mechanics
approach is the Finite Element Method (FEM),
while probably the most common heuristic used for
soft-bodies are the mass-spring systems (MSS),
which were also chosen for this work in order to be
able to achieve faster execution.
As the name implies, the object (its surface only
or the whole volume) modelled by MSS is discre-
tized into a set of point masses (particles) which are
connected by springs. A particle is defined by mass
and position. A spring is defined by stiffness and
damping coefficients, rest length (length of the
spring in rest position) and the two particles it con-
nects. Hooke’s law describes the force acting on the
connected particles by a “spring equation”, while the
movement of each particle is described by usual
Newtonian mechanics. All the particles and spring
parameters do not have any connection to the object
they represent, so it can be difficult to set these pa-
rameters to express the behaviour of the simulated
material correctly.
The MSS methods are popular especially in the
field of cloth modelling and a lot of materials on this
topic can be found for example in a recent survey by
Long et al. (2011). The application of MSS for med-
ical purposes is much less common, mainly due to
the lack of physical accuracy. Nedel and Thalmann
(1998) were one of the first to exploit MSS for mus-
cle simulation. They model only the surface of the
muscle, aligning vertices of the surface triangular
model with the particles of the MSS and its edges
with the springs. They introduce additional angular
springs to limit torsion and also volume loss of the
muscle. This model is used for visualization. Exter-
nal forces yield from the underlying action line
model that approximates all the muscle fibres of a
muscle by one poly-line. While fast to process, an
assumption that all the muscle fibres have the same
length can lead to a wrong force load analysis.
Villard et al. (2008) combined the elastic springs
of MSS with solid parts to create a system which is
still elastic, yet compressible only to a certain extent
and successfully used it to model diaphragm motion.
Hui and Tang (2009) used the MSS to model tendon
GRAPP2014-InternationalConferenceonComputerGraphicsTheoryandApplications
302

motion and deformation. Their system was also
based on the work of Nedel and Thalmann (1998),
using additional flexion and angular springs in order
to contain the torsion and other unwanted defor-
mations.
A recent survey of Lee et al. (2012) compiles a
comprehensive set of various approaches to muscle
modelling, including FEM and MSS based models
and also fast data driven approaches suitable for
non-medical purposes. A reader will find many
references to noteworthy publications there.
2.2 Collision Detection
Detecting collisions of two complex objects in fact
means detecting collisions between two groups of
primitives, i.e. triangles in the most common case of
triangular meshes. As there can be thousands or even
millions of triangles involved in the scene, almost all
collision detection (CD) algorithms include some
pruning phase that limits the number of primitives
that have to be checked in the phase of piecewise
tests.
Methods based on Bounding Volume Hierarchies
(BVH) divide the object into a hierarchical structure
of simple wrapping geometric shapes such as axis
aligned (AABB) or oriented (OBB) bounding boxes,
spheres etc. CD of two objects starts with testing the
root node of the BVH of one object, which bounds
the whole object, with the root of the other. If an
overlap is detected, the following levels of the hier-
archy are traversed and tested until there is no inter-
section detected or until the lowest level is reached.
In the latter case, the primitives stored in the leaf
nodes are subjected to piecewise testing.
A fundamental flaw in the BVH approach in the
context of soft-bodies is that as the object changes
shape, the bounding structure constructed above it
may become invalid. Therefore, before each CD, the
BVH needs to be validated and updated if needed.
There are two common update mechanism – refitting
and rebuilding. Refitting only updates individual
bounding volumes, usually inflating them in order to
accommodate primitives that moved outside their
bounds. When the changes in the shape are too vast
for refitting, the BVH needs to be rebuilt. Levels in
the hierarchical structure are removed bottom-up
until lowest valid level is reached and then the hier-
archy is built again. These operations obviously
need to be fast, which is why the simplest bounding
primitives – usually AABBs – are most commonly
used, even though they might not have such a tight
fit (and therefore less “false positive” overlap tests)
as other shapes used in CD for rigid objects.
Larsson and Akenine-Möller (2006) proposed a
robust BVH solution for CD of deformable and even
breakable objects. Their solution exploits assumed
temporal coherence of subsequent CD steps – the
nodes, that were used in previous CD query are
marked as active and refitted during the update step.
Also, the nodes are validated in that step by compar-
ing the volume of a node to sum of volumes of its
children. If the difference is too large, the children
nodes are destroyed. However, they are not rebuilt
until they are needed.
Mendoza and Sullivan (2006) introduced inter-
ruptible algorithm using BVH for time critical CD
between soft-bodies. Tang et al. (2009) created a
parallel BVH-based algorithm for continuous CD.
They introduced several advanced pruning concepts
that allowed them to achieve interactive frame rates
for scenes with several tens of thousands triangles,
showing that BVH can be used efficiently for soft-
body CD.
Other methods suitable for the soft-body vs. soft-
body CD include spatial hashing techniques
(Teschner et al, 2003), (Hoppe and Lefebvre, 2006)
or methods using the layered depth image (Faure et
al, 2008). As the BVH methods were chosen for the
solution, details about these methods will not be
discussed here.
3 SOLUTION DESIGN
This chapter will describe the devised solution for
muscle simulation. The assignment can be defined
as follows. First, the solution must correctly deform
each muscle in a given group of muscles based on
the interaction with their surroundings (including
other muscles) while undergoing a predefined mo-
tion animation. Interpenetration of individual objects
must be prevented and the results must be presented
in an interactive rate for a single area of interest (e.g.
one lower limb or one upper limb etc.).
3.1 Overall Pipeline
The input data consist of triangular meshes of the
muscles and bones, polylines representing muscle
fibres and transformation matrices for the bones for
each time step, i.e. the motion animation. The user
interface was designed in such a way that enables
rendering an arbitrary point on the timeline (time
point), i.e. it is not assumed that the animation will
be temporarily continuous. For this reason, whenev-
er a given time point is processed, the rest pose of
the muscles and bones is the starting point – all
DeformableMuscleModelsforMotionSimulation
303
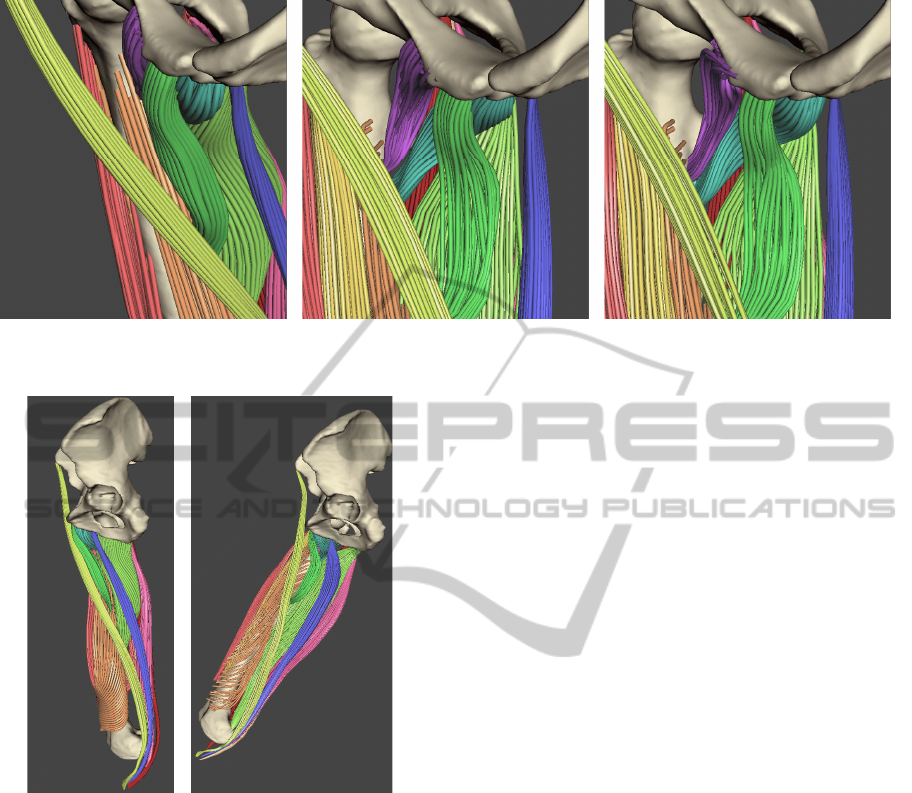
Figure 1: A detailed visualization of muscle fibres in the rest position (left), after rigid transformation to the current position
(middle) and after stabilization of the MSS (right).
Figure 2: Overview of the bone positions for the situation
depicted on Figure 1. Left is the rest position (related to
Figure 1-left) and right is the position during walking
(related to Figure 1-middle and right).
transformations are related to this position.
The first step is pre-processing, which converts
the muscle fibre polylines into particles and springs.
This is controlled by a single parameter, the number
of particles per fibre N. The fibres are divided into
N-1 uniform intervals, but instead of placing the
particles as the end points of those intervals, their
position is randomized within each interval. This is
done because when N is small and the fibres of the
muscle are roughly the same length, which is often
the case, the particles placed without randomization
tend to create clusters with large gaps in between
them and that negatively affects the CD precision
(the CD uses the particles as an approximation of the
muscle surface, therefore they should be scattered
along the surface as much as possible, details in
section 3.3).
Once the particles are created, the spring inter-
connections between them are generated using a
user-selected pattern (see 3.2 for list of implemented
patterns) and then all this is stored as simple arrays,
floating point for the particle positions and pair of
integer indices for each spring. Some other minor
structures are created during the pre-processing as
well, such as associations of particles with bones
closest to them, markings of boundary particles etc.
(see section 3.2 for details). This representation is
stored and reused through different time points un-
less the user changes some key parameters.
To process a given time point of the motion, the
transformation matrices are first applied to the
bones. The muscles (the triangular surfaces as well
as the particles) are transformed as well, using an
interpolated transformation matrix of the nearest
bones. After this basic rigid transformation, the soft-
body simulation takes place.
The simulation is an iterative process consisting
of two major parts - MSS update followed by colli-
sion handling. During the MSS update, new posi-
tions of particles are computed based on the forces
acting in the MSS. These new positions will likely
introduce some collisions with neighbouring objects
and handling those collisions in return adds some
new forces into the system. In an ideal situation,
after iterating this loop a finite number of times, the
MSS should reach an equilibrium state where no
particles move anymore. Then the final shape of the
muscles is ready and can be displayed.
However, an absolute equilibrium is obviously
practically impossible to achieve during computer
simulation. There are three possibilities of how to
end the simulation loop, each equally simple to im-
GRAPP2014-InternationalConferenceonComputerGraphicsTheoryandApplications
304

plement. In order to achieve best fidelity of the out-
put, an average displacement of the particles can be
monitored and when it falls beneath a given thresh-
old, the state is considered final. Or, if the rendering
time is more critical, a given time window, e.g.
based on the desired frame rate, can be assigned to
the computation and the output is produced immedi-
ately after this window is depleted. A compromise is
to set a fixed number of iterations.
After the final particle positions – which define
the final shape – are computed, the surface model is
updated (see 3.2) and visualized. Figures 1 and 2
show the main stages of the process on an example
of a right leg performing a common walking step.
As there are generally many time points in the
animation, the created result is discarded after the
user moves onto another time point. This unfortu-
nately slows the execution when a whole continuous
animation is wanted, as each time point is handled
separately, without exploiting the temporal coher-
ence in any way. For example, the rigid transfor-
mations from rest position to the current position
could be omitted altogether. Moreover, as it can be
expected that there will be only relatively small
changes in consecutive frames, a much smaller
number of iterations of the simulation loop would be
required to get to a plausible state in consecutive
time points. Nevertheless, the application for which
this method was designed required arbitrary time
point changes. Should the requirements change
however, the algorithm would not be difficult to
modify.
3.2 Muscle Model
There are two muscle models – the triangular sur-
face and the other is the muscle fibre model. As was
mentioned before, the key model used in the work-
load analysis is the fibre one, the surface model is
used only for the visualization. MSS are used to
represent the muscles. The MSS solver was already
implemented in the target framework, more details
about it can be found in Zelený (2011).
The solver is computing particle positions in eve-
ry time step of the simulation loop. As the particles
interconnected by springs along the muscle fibre
actually represent this fibre, it is straightforward to
obtain its new shape after the simulation ends since
it is still the same polyline, only its vertices have
different positions. However, it is not as easy to
obtain the new shape of the muscle surface. For this
reason, some relation between the particles and the
triangular model has to be established. We use mean
value coordinates (MVC) for this purpose, described
by Ju et al. (2005) as follows:
The mean value interpolation interpolates a giv-
en function f(x) defined on a closed surface by a
function g(v), v
∈R
3
by projecting a point p(x) of the
closed surface on a unit sphere centered at v. Then the
function value associated with p(x) is weighted by
w=[p(x)-v]
-1
and integrated over the sphere. To ensure
affine invariance, the result is divided by the weight
function integrated over the sphere S. Equation (1)
shows the result:
dSvxw
dSxfvxw
vg
),(
)(),(
)(
(1)
The authors then continue to derive the MVC for
closed polygons and mainly triangular meshes.
Equation (2) computes the weights for point v in
regards to a given triangle with vertices p
i
, i = 1, 2,
3. n
i
are normal vectors of the three triangles vq
i-1
q
i+1
where q
i
are the vertices of a spherical triangle con-
structed by projecting the original triangle p
1
p
2
p
3
on
the unit sphere. m is the “mean vector” which is an
integral of the outward normal vector over the spher-
ical triangle surface (which is not difficult to evalu-
ate using a few trigonometric functions, see Ju et al.
(2005) for details). Weights are computed this way
for all triangles and summed analogically as in equa-
tion (1) (the integrals are replaced by sums over all
the primitives) to obtain the MVC.
)( vpn
mn
w
ii
i
i
(2)
In our case, the boundary (located on outmost fibres)
particles are triangulated and used as the source
mesh, i.e. each particle is the p(x) acting in equations
(1) and (2). Each vertex of the muscle surface mesh
is then treated as the target point (v in (1) and (2))
and its MVC are computed and stored as a pre-
processing. After the particles move during the soft-
body simulation, shape of the surface model is up-
dated simply by recalculating position of each vertex
as a linear combination of its MVC and the current
particle positions. Figure 3 shows the Gluteus max-
imus and Iliacus muscles, which change their shapes
significantly during the simulated motion – in this
case simple walking. The MVCs are used to deform
the surface from the rest pose to the final pose while
conserving the smoothness of the surface.
To simulate the tendon attachment of each mus-
cle, the particles on ends of the fibres are set as
fixed. The position of fixed particles is not updated
in the soft-body simulation, only during the rigid
transformations.
DeformableMuscleModelsforMotionSimulation
305
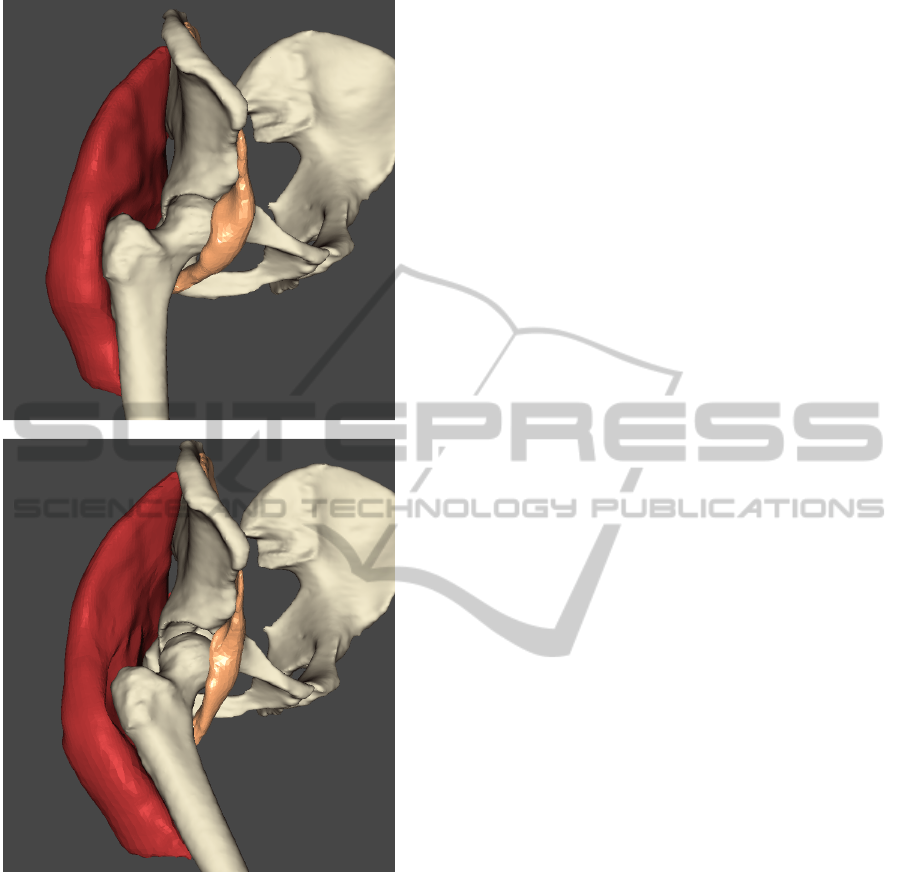
Figure 3: Surfaces of the Gluteus maximus and Iliacus
muscles. Up in rest pose (standing), down in the final pose
(walking).
There are many ways, or templates, of how to
generate the springs connecting the particles. The
more springs there are, the longer the computation
will be, but on the other hand, systems with low
spring count tend to converge to the equilibrium
state slower. Several configurations were tested:
cubic lattice with 6 or 26 springs per particle (the
particles are treated as vertices of a cubic grid with
either only the sides of the cubes being the springs,
or all sides and both face and space diagonals being
the springs); delaunay tetrahedralization (springs are
generated as edges of tetrahedrons in a delaunay
tetrahedalization of the particles); and N nearest
neighbours (each particle is connected with a user
defined number of closest particles). The impact of
the choice of the layout is discussed in Section 4.1.
3.3 Collision Handling
Collision handling is responsible for ensuring that no
objects penetrate each other. However, as section 3.2
established, the surfaces of the muscles are not in-
volved in the simulation at all. That means that one
would have to update the surface mesh in each step
of the simulation loop, detect collisions, propagate
the response onto the particles via the MVC and
continue with the simulation. This would significant-
ly increase the computation time. Another problem
is that the surfaces of individual muscles can actual-
ly intersect even in the initial position. That is not an
error in the data – some muscles can, and do, inter-
weave each other, which is easily simulated by using
the fibre model, but difficult when using the encap-
sulating surface.
To bypass these problems, the particle model it-
self is used for the collision detection (CD) instead.
Obviously, the particles represent point masses and
therefore do not have any volume. Also, they do not
necessarily trace the surface of the muscle. To
change that, the particles are thought of as spheres
with a given non-zero radius. Interpenetrations be-
tween these sets of spheres for different muscles can
then be detected and removed. At the same time
their position coincides with the particle positions
and therefore the collision response consists of noth-
ing more than updating the positions of the particles.
Another pleasing by-product of this design is simpli-
fication of the CD itself, as detecting and resolving
collisions of set of spheres is generally simpler than
in the case of triangle meshes – sphere vs. sphere
collision test is a simple comparison of their distance
and the sum of their radii. Also, only the particles
that are on the “boundary” fibres, i.e. fibres closest
to the surface, have to be accounted for – the internal
particles will not collide with other objects due to
the internal forces of the MSS.
The following heuristic has been used to obtain
the radius for each sphere. The closest particle to
each vertex of the surface mesh is found. Note that
one particle can be the closest one to several verti-
ces. To make a compromise between covering
enough of the surface of the muscle and the least of
the outside space, the radius of each sphere is set to
an average of the distances between the centre of the
particle and the associated vertices. Then the CD
mechanism is employed to detect particles that inter-
GRAPP2014-InternationalConferenceonComputerGraphicsTheoryandApplications
306
sect and their radii are decreased so that the intersec-
tion are removed, which effectively removes some
very large particles that could have been generated.
To increase the coverage of the surface without
increasing the volume excessively, more particles
per fibre can be used. However, that comes at a price
of higher memory and time demands.
For collision detection between the muscles and
bones, the same mechanism is used. The bones are
also converted to a set of spheres that approximates
their surface and then the muscle vs. bone CD is
processed in the same way as muscle vs. muscle.
The position of the spheres representing the bone
can be arbitrary (unlike in the case of muscles) and
therefore the spheres can trace the surface more
closely without taking up too much excess space.
The CD itself uses the BVH mechanism based on
the method by Larsson and Akenine-Möller (2006),
using AABBs, subdivided into even octants (i.e. the
division lines always passes through the midpoint of
the parent box). This method was chosen over the
others mentioned in Section 2.2, due to its universal-
ity and good performance documented in the afore-
mentioned work. Simple AABBs are used, because
they are faster to update as the object changes shape.
The bounding boxes are subdivided into octants in
each level of the subdivision. The division lines
always pass through the midpoint of the parent box.
Whenever new object is added to the scene, the
bounding box is constructed for it only on the parent
level, i.e. without any subdivision.
The rebuilding proposed by the original authors
(see 2.2) is not used. After testing, it has been found
out, that the proposed rules for determining whether
to rebuild part of the hierarchy does not work well in
this case. The CD is on average 5% faster when only
refitting is used. That is actually not very surprising,
as it can be expected that the shape of the muscles
will not change drastically during one simulation
step, when the particles only head toward the equi-
librium state. Also, the bounding boxes of the bones
obviously never need to be updated, because they do
not move (not in the span of one simulation step) or
change shape.
Each muscle in the scene is then tested against
each other muscle and also each bone. The BVHs of
the objects are traversed and subdivided only when
needed. The maximal number of recursion steps as
well as the minimal number of primitive per node
are either specified by a user or their default values,
empirically determined to be 8 and 10, respectively,
are used. Once either of the limits is reached, and
there is still a collision detected between the nodes, a
pair of lists containing the primitives in the two
colliding nodes is outputted and the piecewise test of
the primitives in these lists is done.
When the piecewise test detects a collision, the
difference between the distance of the two particles
and the sum of their radii is computed. Then each
particle is moved in an opposite direction to each
other by half of this distance, or eventually, if a
fixed particle is involved, the unfixed particle is
moved by the whole distance (spheres that represent
bones are treated as fixed particles). This way the
particles end up just touching each other. To take
into account that a given particle can collide with
multiple other particles, the displacement vectors are
stored and accumulated for each particle and only
after all test have been made, their superposition is
applied to the initial particle positions.
After all tests are finished, the results are applied
(i.e. the accumulated displacement vectors are used
to update the positions) and a new iteration may
begin. The particles that collided in a given time step
are treated as stopped – they are assigned a zero
velocity. This ensures that they do not rebound after
collision, which is natural for the kind of elastic
behaviour that is being simulated.
4 EXPERIMENTS
Our approach was implemented in C++ using the
VTK library (http://www.vtk.org) and integrated in a
MAF based (http://www.openmaf.org) application-
created for the VPHOP project mentioned earlier.
Figure 4: Overview of the used dataset.
DeformableMuscleModelsforMotionSimulation
307
Tests were done on a PC with Intel Core i7-3770
(3,4GHz, 4 physical cores with hyper-threading)
CPU, 8GB RAM, MS Windows 7 64-bit OS.
One data set was used for all the tests. It consists
of MRI footage of pelvis and legs, fused with a mo-
tion capture data of a walking human. A total of
twenty three muscles are available for testing in the
data set, ten of which are on the pelvis and thirteen
on the right thigh. The visualization of this dataset is
on Figure 4, with muscles rendered as surfaces.
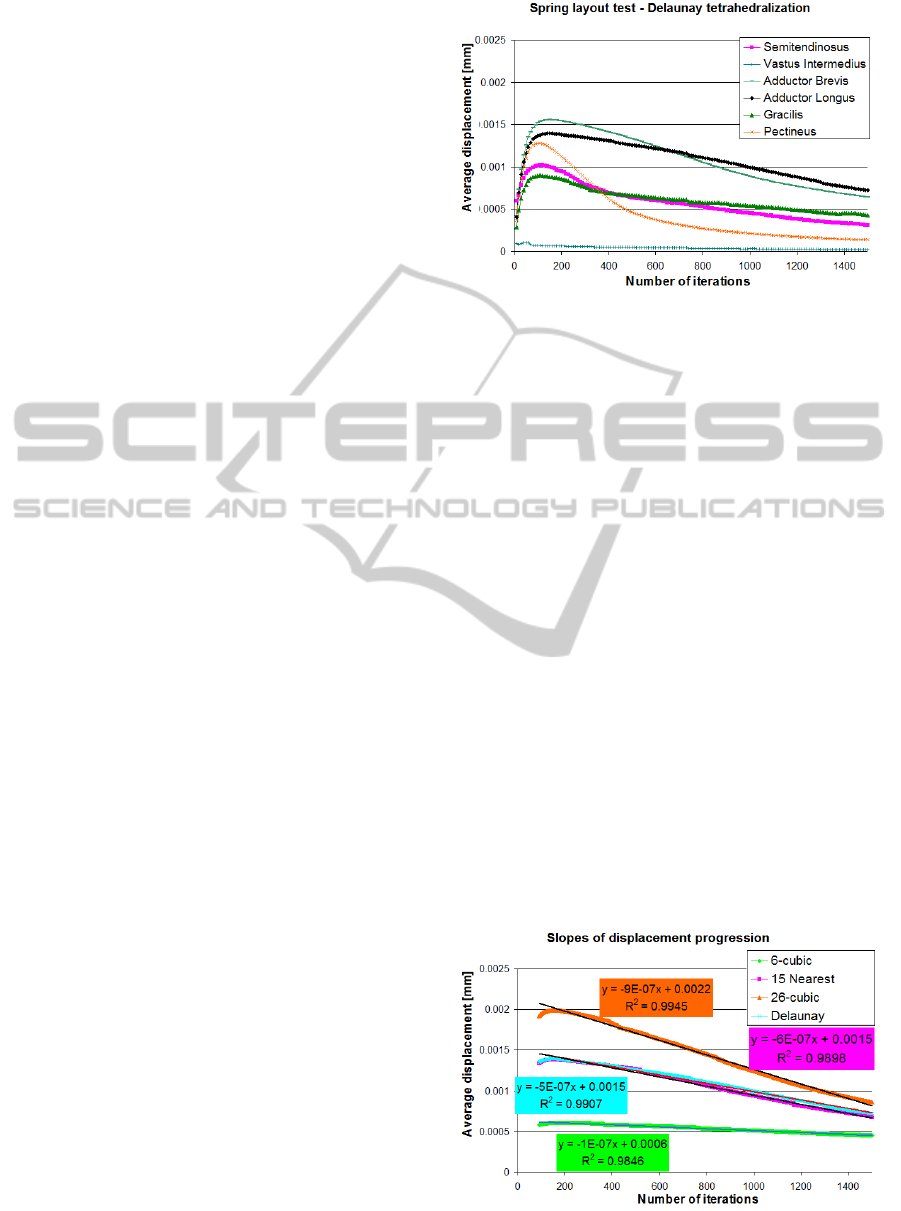
4.1 Spring Layout
The number of iterations needed to achieve the final
shape of the soft-body affects the computational
time the most. The spring layout has a significant
impact on this number, therefore several layouts
were tested. The test processed 1500 iterations of the
simulation for each tested method, measuring the
average displacement of particles, i.e. the difference
in position of each particle between two successive
iterations. In an ideal state, the particles would not
move at all once the equilibrium position is
achieved. However, this is unlikely to happen in the
simulation. Rather than zero displacement, an oscil-
lation of the displacement values is a sign of the
final state (the positions will oscillate due to the
continuous collisions between adjacent particles).
The tested layouts were: Cubic lattice models
with 6 and 26 springs per particle, Delaunay tetrahe-
dralization (DT) and 15 nearest neighbours (see
section 3.1). The times for those methods were
111.12s, 129.6s, 124.19s and 121.37s respectively,
but please note that these times are listed only for
comparison between different spring layouts and do
not reflect the performance of the final solution – see
section 4.3 for a complete analysis of time consump-
tion. The thirteen thigh muscles were used for these
tests.
Figure 5 shows the absolute displacements for
six of the tested muscles (for better clarity of the
chart, some muscles were omitted) for the DT. The
displacement rises in the beginning as the particles
start to move faster due to the forces introduced into
the system by the initial rigid transformations. As
they reach the proximity of their final positions, their
movement slows down (after around 80 iterations),
ideally stopping entirely after more iterations.
The 26-cubic lattice and 15 nearest neighbours
have fairly similar progression as the DT on Figure
5, while the 6-cubic lattice displays almost an order
lower differences. However, note that the absolute
value of displacement measures only the differences
between consecutive iterations, not how close are
Figure 5: Absolute displacements for several muscles
simulated with the DT spring layout.
these positions to the final state. It is desirable for
the differences to actually be as high as possible.
That would mean that the model is moving fast
towards the final configuration of positions.
Figure 6 depicts the displacements for the Ad-
ductor Longus muscle (as Figure 5 documents, the
progression is quite similar for all the muscles,
therefore the choice of test subject is not important)
using four different layouts and including a linear
regression of the trend for each layout. Several first
iterations were not accounted for, because it is the
later “stabilization” phase where the differences
between individual layouts have the highest impact.
It is apparent that the 6-cubic lattice layout con-
verges at the slowest rate, while the 26-cubic is the
fastest. Even though the 6-cubic layout is the fastest
per iterations, much more iterations will be needed
in order for the model to reach the equilibrium and
therefore the execution will be the slowest. The
remaining two methods offer a reasonable compro-
mise. Various count of neighbours in the nearest
neighbour method were tested, revealing almost
strictly linear relation between the number of springs
and convergence speed.
Figure 6: Slopes of particle position displacement for the
Adductor Longus muscle.
GRAPP2014-InternationalConferenceonComputerGraphicsTheoryandApplications
308

The 6-cubic lattice is clearly the least suitable
option. Although 26-cubic lattice provides fastest
convergence, the nearest neighbour or DT layouts
also provide fast convergence while having slightly
lesser memory and computation time demands.
Moreover, if the nearest neighbour is used, the num-
ber of neighbours can be provided to the user as an
additional parameter to control the model (also note
that when using 26 neighbours, the behaviour will be
very similar to the 26-cubic). Hence, the nearest
neighbour is the suggested solution.
4.2 Deformation Quality
To compare the results of various deformation
methods, volume preservation is often used. It is
disputable whether this is a relevant metric, since the
assumption that a muscle retains its volume does not
hold in general. Nevertheless, it is one of few meas-
urable characteristics of different methods and can
reveal some relations between them. The error in
muscle volume preservation was on average 2.71%
for the used dataset. Although larger than the other,
more precise method that was available as part of the
project solution (which averaged on 0.08%), up to
6% errors are considered to be acceptable and there-
fore the presented method is usable. Moreover, by
using more fibres for the model and a larger stiffness
coefficient for the springs, the volume loss should
decrease.
More relevant evaluation method is a visual
check and comparison with real data by an expert.
Because it is almost impossible to obtain real MRI
images of the patient in different poses, medical
literature must suffice as the data in this case. The
implemented solution was handed over to a partner
facility (Istituto ortopedico Rizzoli, Italy,
http://www.ior.it) to do such evaluation test. The key
observation was that our fibre model shows “reason-
able consistency with the behaviour reported in
literature”.
The partner further investigated changes in fibre
length during motion. In general, the method per-
formed well, emulating behaviour described in med-
ical literature closely. The only complication arose
when the muscle was shortened significantly, as the
MSS-driven model tend to buckle as it was resisting
the shortening. Lower stiffness coefficients remove
this problem, but, as mentioned before, lower stiff-
ness also tend to result in higher volume error. To
conclude, in order to satisfy both the demand for low
volume error and the ability to correctly model
shortening, the stiffness coefficient must be small
and the fibre count (and therefore particle count)
high. Of course, higher particle count means slower
execution, so a balance between those parameters
has to be found. Nevertheless, the partner facility
found the method suitable for the target application.
4.3 Overall Time Performance
In order to evaluate the performance of the method,
computation times of individual steps of the simula-
tion were measured. 500 iterations of the simulation
loop were set for the test. This number was chosen
solely to obtain significant volume data for the time
measurements. In real application, less or more itera-
tions might be needed, depending on the desired
precision. Note however, that for the tested models,
500 iterations were enough to reach a state in which
the MSS start oscillating around the same values
even for the lowest particle count. Only the 13 thigh
muscles were used for this test.
Four different particle resolutions (i.e. numbers
of particles per fibre) were used for the evaluation –
20, 40, 60 and 80. While twenty particles per fibre
are sufficient to obtain acceptable output (i.e. the
model does not diverge to unnatural shapes), higher
resolution might offer better ratio between conver-
gence speed and output quality. The number of fi-
bres per muscle was always 64. While the speed of
the MSS simulation depends on the total number of
particles, the speed of CD depends only on the num-
ber of the boundary particles, as only those are in-
volved in CD. The total number of particles of the
muscles / the number of boundary particles for indi-
vidual resolution are: 1344 / 660 for resolution 20,
2624 / 1220 for 40, 3904 / 1780 for 60 and 5184 /
2340 for 80. There were three bones significant for
the used set of muscles. The same resolution (num-
ber of spheres representing the bone) was used in all
cases: 8527 spheres for the pelvis, 1568 for femur
(thigh) and 1709 for tibia (shank).
Table 1 shows the result of the test. The “MSS”
line contains the time of simulation of the MSS (the
26-cubic lattice layout was used). “Muscle CD” line
contains the time of CD of muscles vs. muscles and
the “Bones CD” contains the time of muscles vs.
bones CD. The times do not include any pre-
processing. Lastly, because the results are also af-
fected by the pose of the model (i.e. there might be
more collisions in one position than some other), the
provided times are actually an average of five differ-
ent poses of the model.
The used MSS solver is only a basic, non-
parallel implementation which can be improved on
to achieve better results. The author of the solver,
Zelený (2011), also implemented a GPU version
DeformableMuscleModelsforMotionSimulation
309

Table 1: Computation times in seconds for individual parts
of the method after 500 simulation iterations.
Resolution
Part
20 40 60 80
MSS 18.66 39.97 60.53 80.70
Muscle CD 8.15 13.50 24.00 29.53
Bones CD 30.03 37.43 56.03 67.10
TOTAL 56.84 90.9 140.56 177.33
obtaining on average 5-8x speed-up, depending on
the number of particles. However, his latest imple-
mentation cannot handle large systems due to
memory issues and would have to be modified first.
As this work was not finished yet, the CPU solver
was used here instead. Also note that the higher
computation time of CD between bones and muscles
than muscles vs. muscles is expected due to the
higher overall resolution of the bones (see above).
Table 1 shows a linear relation between the
number of particles and CD time consumption,
which is rather disappointing, as the relation should
be, in ideal case, logarithmic due to the hierarchical
structure. On average, 76% of the CD computation
time is the hierarchy traversal and 24% the piece-
wise testing. In the time counted as the piecewise
testing there is also accounted the collision response,
i.e. computing the vectors by which the spheres are
moved (see section 3.3), therefore the traversal is
considered the bottleneck.
Possible explanation of the slower-than-expected
performance is the loose fit of AABBs. As all the
objects occupy more or less the same space (they are
wrapped around the same bone), the first levels of
the hierarchy of two muscles will always intersect,
resulting in many “dead end” traversals. However,
even if for example OBBs were used instead, the
situation would not improve much as there would
still be significant overlap of the bounding volumes
due to the nature of the data.
5 CONCLUSIONS
A mass-spring model designed for representing
muscles in a musculoskeletal model was presented.
The mass-points are obtained by sampling the fibres
of the muscle. Three different ways of connecting
the particles by springs were tested. The importance
of the layout itself was found to be only marginal,
while the number of springs per particle influences
the convergence speed the most.
A collision detection and response mechanism
was designed, implemented and tested. The mecha-
nism employs bounding volume hierarchies to en-
hance the speed, the particles of the mass-spring
system are utilized as spheres of various radii ap-
proximating the surface of each object. This does not
only speed up CD tests, but also bypasses the need
to propagate changes of the shape between the sur-
face model and the mass-spring model in each itera-
tion of the simulation.
The proposed solution was evaluated by medical
experts and deemed as suitable for the purpose of
quick coarse simulations of moving human patient
and subsequent analysis of muscle deformation.
Other methods for collision detection should be
investigated further as the future work. It is the bot-
tleneck of the solution and the current mechanism
does not perform as well as expected. However, the
given task is quite complex, therefore there is no
guarantee that other algorithms will perform better.
ACKNOWLEDGEMENTS
This work has been supported by the Information
Society Technologies Programme of the European
Commission under the project VPHOP (FP7-ICT-
223865), the European Regional Development Fund
(ERDF) – project NTIS (New Technologies for
Information Society), European Centre of Excel-
lence, CZ.1.05/1.1.00/0.2.0090 and by the project
SGS-2013-029 – Advanced Computing and Infor-
mation Systems.
REFERENCES
Faure, F., Barbier, S., Allard, J., Falipou, F., 2008. Image-
based collision detection and response between arbi-
trary volume objects. In Proceedings of the 2008 ACM
SIGGRAPH/Eurographics Symposium on Computer
Animation (SCA '08). Eurographics Association, Aire-
la-Ville, Switzerland, Switzerland, 155-162.
Ju, T., Schaefer, S., Warren, J., 2005. Mean value coordi-
nates for closed triangular meshes. In ACM SIG-
GRAPH 2005 Papers (SIGGRAPH '05), Markus
Gross (Ed.). ACM, New York, NY, USA, 561-566.
Lefebvre, S., Hoppe, H., 2006. Perfect spatial hashing. In
ACM SIGGRAPH 2006 Papers (SIGGRAPH '06).
ACM, New York, NY, USA, 579-588.
Larsson, T., Akenine-Möller, T., 2006. A dynamic bound-
ing volume hierarchy for generalized collision detec-
tion. Computer Graphics 30, 3. Pergamon Press, Inc.
Elmsford, NY, USA, 450-459.
Lee, D., Glueck, M., Khan, A., Fiume, E., Jackson, K,
2012. Modeling and Simulation of Skeletal Muscle for
GRAPP2014-InternationalConferenceonComputerGraphicsTheoryandApplications
310

Computer Graphics: A Survey. Foundations and
Trends in Computer Graphics and Vision, 7, 4. Now
Publishers Inc. Hanover, MA, USA 229-276.
Lee, S.-H., Sifakis, E., Terzopoulos, D., 2009. Compre-
hensive biomechanical modeling and simulation of the
upper body. ACM Trans. Graph. 28, 4, Article 99, 17
pages.
Long, J., Burns, K., Yang, J., 2011. Cloth modeling and
simulation: a literature survey. In Proceedings of the
Third international conference on Digital human
modeling (ICDHM'11), Vincent G. Duffy (Ed.).
Springer-Verlag, Berlin, Heidelberg, 312-320.
Mendoza, C., O'Sullivan, C., 2006. Interruptible collision
detection for deformable objects. Comp. Graphics 30,
3. Pergamon Press, Inc. Elmsford, NY, USA 432-438.
Nedel, L. P., Thalmann, D., 1998. Real Time Muscle
Deformations using Mass-Spring Systems. In Pro-
ceedings of the Computer Graphics International 1998
(CGI '98). IEEE Computer Society, Washington, DC,
USA, 156-.
Tang, M., Manocha, D., Tong, R., 2009. Multi-core colli-
sion detection between deformable models. In 2009
SIAM/ACM Joint Conference on Geometric and Phys-
ical Modeling (SPM '09). ACM, New York, NY,
USA, 355-360.
Tang, Y.-M., Hui, K.-C., 2009. Simulating tendon motion
with axial mass-spring system. Comp. Graphics 33, 2.
Pergamon Press, Inc. Elmsford, NY, USA, 162-172.
Teschner, M., Heidelberger, B., Mueller, M., Pomeranets,
D., Gross, M., 2003. Optimized Spatial Hashing for
Collision Detection of Deformable Objects. In Pro-
ceedings of Vision, Modeling, Visualization VMV'03.
Munich, Germany, 47-54.
Villard, P.-F., Bourne, W., Bello, F., 2008. Modelling
Organ Deformation Using Mass-Springs and Tension-
al Integrity. In Proceedings of the 4th international
symposium on Biomedical Simulation (ISBMS '08),
Fernando Bello and P. J. Edwards (Eds.). Springer-
Verlag, Berlin, Heidelberg, 221-226.
Zelený I.: Vzájemná transformace 3D objektů reprezento-
vaných trojúhelníkovým povrchem [Morphing of 3D
objects represented by triangular surface]. Diploma
thesis, University of West Bohemia, Faculty of applied
sciences, 2011. (Available only in Czech language).
DeformableMuscleModelsforMotionSimulation
311
