
Bark Recognition to Improve Leaf-based Classification in Didactic Tree
Species Identification
Sarah Bertrand
1
, Guillaume Cerutti
2
and Laure Tougne
1
1
Univ. Lyon, Lyon 2, LIRIS, F-69676 Lyon, France
2
INRIA, Virtual Plants INRIA Team, Montpellier, France
Keywords:
Bark, Leaf, Tree Recognition, Smart-phone.
Abstract:
In this paper, we propose a botanical approach for tree species classification through automatic bark analysis.
The proposed method is based on specific descriptors inspired by the characterization keys used by botanists,
from visual bark texture criteria. The descriptors and the recognition system are developed in order to run on
a mobile device, without any network access. Our obtained results show a similar rate when compared to the
state of the art in tree species identification from bark images with a small feature vector. Furthermore, we
also demonstrate that the consideration of the bark identification significantly improves the performance of
tree classification based on leaf only.
1 INTRODUCTION
Nowadays, the growth of urbanization and techno-
logy has reduced the knowledge and uses of plants
by humans. However, the environment and its na-
tural resources are raising a growing concern. The
ability to identify the species of plants seems essen-
tial in the understanding of our green environment.
Thereby, providing an inexperienced person, who had
no knowledge in botany, with a tool to identify the
plants that surround him would be a great advance.
The portion of the population using a smart-
phone, incorporating a camera, a GPS, etc. grows in-
creasingly. These phones allow furthermore to down-
load additional applications with other functions than
communication. These smart-phones are ideal mate-
rials for applications related to image analysis.
From these two facts, let us consider the case
where a person, wandering through a forest or a city
park, wishes to recognize a species of tree. He then
takes a picture of the different organs of the plant
(leaf, bark, flower) using his mobile phone. By
means of interactions between the smart-phone (via
the touch screen) and the user, and of course of treat-
ments and image processing, the system would return
a short list of species that may correspond to the plant
the user seeks to identify, coupled with a confidence
rate.
Applications for smart-phone have been de-
veloped to address this subject, such as Leaf Snap
(Kumar et al., 2012), Pl@ntNet (Go
¨
eau et al., 2013a),
or Folia (Cerutti et al., 2013). Furthermore, plant re-
cognition has recently been the subject of research in
the field of image processing. Challenges have been
organized especially the ImageCLEF Challenge since
2011 (Go
¨
eau et al., 2011).
Even though the leaves are the most used organs
for tree classification in the applications, as they are
easily observed and described, the other parts of the
tree can help the recognition of tree species. In this
article, we focus on the recognition of tree species
through the bark. We choose to process this part of
the tree as it is always observable throughout the year
(not like flowers or fruits), and can be easily photo-
graphed.
Since 2013, recognition of plants in the Image-
CLEF Challenge spread to other plant parts and not
only leaves as the 2011 and 2012 editions. In these
challenges, the different teams program algorithms in
order to recognize plant species through several im-
ages of the plant. In (Go
¨
eau et al., 2013b) and (Go
¨
eau
et al., 2014) we can see the evolution of the extracted
features used for the identification of the bark. Un-
til 2014, the majority of the algorithms was based
on the detection and characterization of points such
as SIFT (Scale-invariant feature transform) (Lowe,
1999) or SURF (Speeded-Up Robust Features) (Bay
et al., 2006), with color information as well as in-
formation from Fourier transforms or LBP (local bin-
ary pattern) (Ojala et al., 1996), with bag-of-word ap-
Bertrand S., Cerutti G. and Tougne L.
Bark Recognition to Improve Leaf-based Classification in Didactic Tree Species Identification.
DOI: 10.5220/0006108504350442
In Proceedings of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2017), pages 435-442
ISBN: 978-989-758-225-7
Copyright
c
2017 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
435

proaches. Texture descriptors were not used for the
bark recognition as they gave poorer results. Since the
ImageCLEF 2015 edition (Go
¨
eau et al., 2015), there
is a large use of deep learning approaches, not only
for the classification, but also for the features extrac-
tion. The GoogLeNet convolutional neural network
(Szegedy et al., 2015) has been employed by nearly
half of the teams. Only one team used hand-crafted
visual features.
These methods are efficient but first, they need
lots of training images, which is often hard to obtain
in the botanic domain. Secondly, they run as black
boxes that is to say they only give a classification res-
ult without giving the user the possibility to under-
stand the result. But, the work that is proposed in this
article takes part of a pedagogic project the goal of
which is to learn the user to recognize a tree or a shrub
from its organ. For the leaves, a method based on
botanic criteria has already been developed by (Cer-
utti et al., 2013), which allows showing the user the
discriminant criteria during the recognition process.
The presented work aims at pursuing in this way us-
ing features the botanists use to discriminate the bark.
Notice also that as the goal is to do the image treat-
ment on smart-phones, as very often the Internet is
not reachable in the countryside, so we would like a
feature vector with reasonable size and, computation
time of it and classification time in real time that is to
say in a few seconds.
In the following, we will present in part II the con-
struction of the features vector describing a bark im-
age. The next part III will display our different results
on the bark database and the improvement of the pro-
cessing of the bark on the tree species classification
only based on the leaves.
2 BARK ANALYSIS
Our proposed method differs from the state of the
art in the sense that it is based on the extraction of
descriptors similar to those used by botanists from
visual texture criteria. More specifically, the method
simulates the bark recognition described by Michael
Wojtech in his book (Wojtech, 2011). In this ap-
proach, the barks are first classified into different fam-
ilies as shown in Figure 1.
The first row contains photos of characteristic bark
for each families. The second row highlights the dis-
tinctive bark structure of each photo of bark with a
drawing. More drawings can be found in (Wojtech,
2011). After identifying the type of bark, botanists
also take into account the color or other structural ele-
ments in order to recognize the tree species.
Figure 1: Different classes of bark structure based on visual
criteria.
In the following parts, we present the extracted
features to recognize the various types of barks and
then the different tree species.
2.1 Shape of the Bark
In this section, we propose a method to describe pre-
cisely the structure of the bark, not only whether it
has a vertical or horizontal structure. The goal is to
identify the different types of bark (Figure 1), i.e.,
bring out the forms (scales, straps, cracks, etc) of the
bark. For this, at first, we need to extract the edges
of the shapes. For this purpose, we use the Canny al-
gorithm (Canny, 1986). Let us denote by M
H
and M
V
respectively the horizontal and the vertical maps ob-
tained with a horizontal filter F
H
and a vertical filter
F
V
.
F
H
=
−1 −2 −1
0 0 0
1 2 1
and F
V
=
−1 0 1
2 0 2
−1 0 1
(1)
The results of this Canny processing are visible on
Figure 2 for M
H
and on Figure 3 for M
V
.
Figure 2: Map of horizontal edges M
H
obtained from Figure
1.
Figure 3: Map of vertical edges M
V
obtained from Figure
1.
Using the maps of edges images, we can estimate
the dissimilarity between the different families of
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
436
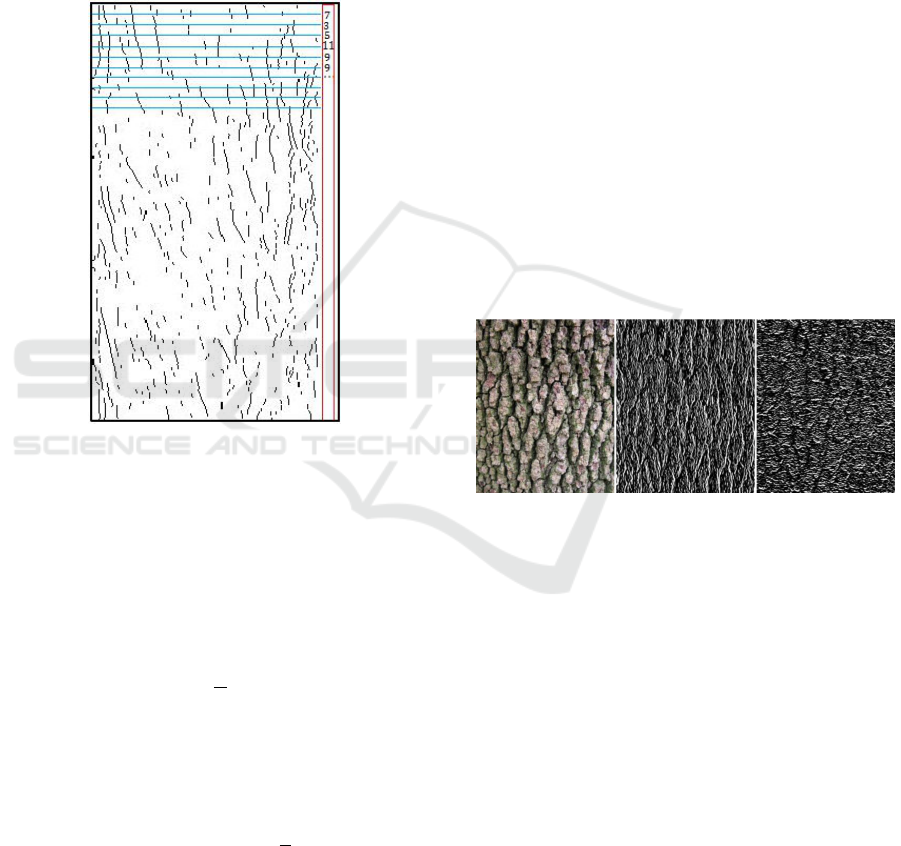
bark. To characterize the distribution of contours, we
propose to place a grid over the maps of edges images,
and to count the number of intersections between the
grid lines and the maps of contours. Specifically, from
M
V
(respectively M
H
), we focus on the intersections
between these contours and horizontal lines (vertical
respectively) grid. Then, for each row (column re-
spectively) grid, we calculate the number of intersec-
tions. This generates a ”vertical word” (respectively
”horizontal word”) (see Figure 4).
Figure 4: Formation of the vertical word.
More formally, let l be the number of rows of
the image, c the number of columns, with λ
l
the fre-
quency of the vertical gridlines and λ
c
the frequency
of the horizontal grid. The higher the frequencies λ
l
and λ
c
, the higher the structure is precisely described.
The ”vertical word” W
V
is the concatenation of the in-
tersections v
l
with l varying from 0 to l with the fre-
quency of the grid λ
c
.
v
l
=
j
c
λ
c
k
∑
c=0
δ
l,c
(2)
with
(
δ
l,c
= 1 if M
V
(λ
l
, λ
c
) = 1
δ
l,c
= 0 else
(3)
and V =
{
v
l
}
0≤l≤
j
l
λ
l
k
(4)
M
H
and M
V
are finally characterized by two
words, respectively W
H
and W
V
, that define the fre-
quency and distribution of the edges. If a word con-
tain mostly zeroes it means that the corresponding ori-
entation is rather smooth. When the ridges of the bark
are practically uninterrupted, the ”vertical word” will
be homogeneous. We will give in the Results section
some elements to explain the choice the discretization
steps in the two directions.
2.2 Bark Orientation
As we can see in Figure 1, barks may have structures
with different orientations, i.e. horizontal, horizontal
and/or vertical, or just be smooth. The orientation of
the bark is a recognition criterion. The image filter-
ing by Gabor wavelet can distinguish the direction
and frequency of the structure of the bark. Accord-
ing to Huang (Huang et al., 2006), only 6 orienta-
tions and 4 scales for the sinusoid forming the Gabor
wavelet are sufficient to analyse the bark. However,
in the case of our application, the distance between
the smart-phone and the tree trunk may vary. Our
method must be scale independent as we can not con-
trol the camerawork. In addition, we want to determ-
ine, initially, whether the structure of the bark is ho-
rizontal or vertical, so only two orientations appears
adequate (0
◦
and 90
◦
). Figure 5 shows an example of
bark filtered by two identical Gabor wavelets except
for the orientation.
Figure 5: Results of 0
◦
orientation Gabor filtering (center)
and 90
◦
(right) of the initial image (left).
In order to have a characteristic vector of each
bark which is independent of the shooting distance,
we average four filtered images by Gabor wavelets
with four different sinusoidal scales. The scales are
adjusted in order to highlight the horizontal and ver-
tical ridges of the bark, similarly to Figure 1.
The feature vector G reflecting the orientation of
the structure using Gabor filters consists of the mean
and the standard deviation of the filtered image with
an orientation of 0
◦
, σ
0
and µ
0
, and the standard devi-
ation image filtered at 90
◦
, σ
90
. We considered in-
cluding also the average of images at 90
◦
, but this
feature appeared as not discriminant, so we decided
to leave it apart, ending up with a simple feature vec-
tor of dimension 3: G : [σ
0
µ
0
σ
90
] . For the moment,
our features vector is composed of this Gabor vec-
tor and of W
H
and W
V
. We will show in the Results
section the positive contribution of the Gabor vector
concatenated with the two words W
H
and W
V
.
Bark Recognition to Improve Leaf-based Classification in Didactic Tree Species Identification
437

2.3 Bark Color
A second discriminant aspect for tree bark recog-
nition used by botanists is the color. Colorime-
tric descriptors were used in the challenge Image
CLEF 2013 by the teams showing the best results
(Bakic et al., 2013) (Nakayama, 2013). Several color
spaces were tested. We favored color spaces where
color channels were independent of brightness as the
descriptor should not be influenced by the presence of
shadows or by the changing of illumination that can
occur over the day and the year. The L* a* b* color
space seemed appropriate because of the channel a*
coding the red/green opposition and as we know the
bark of trees have brown, red, green, or gray shades.
However, the hue channel (H) of the HSV color space
was chosen because it gives us better classification
results. It is because the hue channel can also man-
age the case of yellowish bark. The hue channel al-
lows to cover the whole range possible of bark color
with a single channel with 256 bins, contrary to the
concatenation of the a* and b* channels that would
have given a doubled size color feature vector. As a
reminder, we try to have a reasonable size feature vec-
tor. Figure 6 shows the hue histogram for some bark
images.
Figure 6: Hue histogram for different barks.
Finally, the feature vector used for bark classific-
ation F
b
therefore consists in the concatenation of the
vector of Gabor G, W
H
and W
V
and the hue histogram
H.
3 RESULTS
3.1 Bark Classification
For the bark classification, we have a database of 2559
bark image belonging to 101 tree species from the
data bank Pl@ntView
1
. Pictures of bark have the
same orientation as the tree (portrait mode). In our
practical application, we have re-sized these images
in order that the picture contain only the texture of
the bark, not the background, and no other elements
such as branch, leaves, or lichen. As a matter of
fact, we can make the assumption that the interface
of the application can guide the user to take such a
king of photos. However, we have kept the bark im-
ages which have a gradient of light or shadows (see
Figure7). This allows us to have an algorithm which
should work with images shot in non-optimal condi-
tions by beginners. We may also note that the number
of bark images is not the same for all the species (20
in average, but can go up to 100 for some species and
2 or 3 for others). The low number of samples for
some species makes it difficult to train a performing
classifier.
Figure 7: Bark of Acer Pseudoplatanus with homogeneous
luminosity, gradient of light or shadows.
As expressed in the introduction, we use a SVM
in order to classify the feature vector. The kernel
of the SVM is a Radial Basis Function (RBF) kernel
(Vapnik, 1999). We use a 1-vs-all algorithm, i.e. we
have one binary SVM for each species, with the class
species S versus the class non-S species. For each
query bark, we compute the feature vector then we
give this vector to all the 101 SVM classifiers. After-
wards the result is the species which the correspond-
ing classifier gives the higher probability of affiliation
1
http://publish.plantnet-project.org/project/plantclef
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
438
to the species S. On a Macbook Pro dating from 2011,
with a processor 2.7 GHz IntelCore i7 and a memory
4 GB 1333 MHz, we get a classification result in less
than a second. As the last smart-phones have on aver-
age a processor of 2 GHz and quad-core, that make us
believe that our method could run on these devices.
Train is performed on half of the database, and
tests are performed on the second half. We obtain
a classification rate equals to 30,7%. If we observe
the results of the plant task of the LifeCLEF chal-
lenges (Go
¨
eau et al., 2015), plant recognition via the
stem gives the worst classification rate compared to
the other other organs. The state of the art give around
30% of good identification. So, with our method, we
achieve the same level with a quite short feature vec-
tor and without heavy classification process.
In the case of a smart-phone application, we can
propose to the user a list of tree species which most
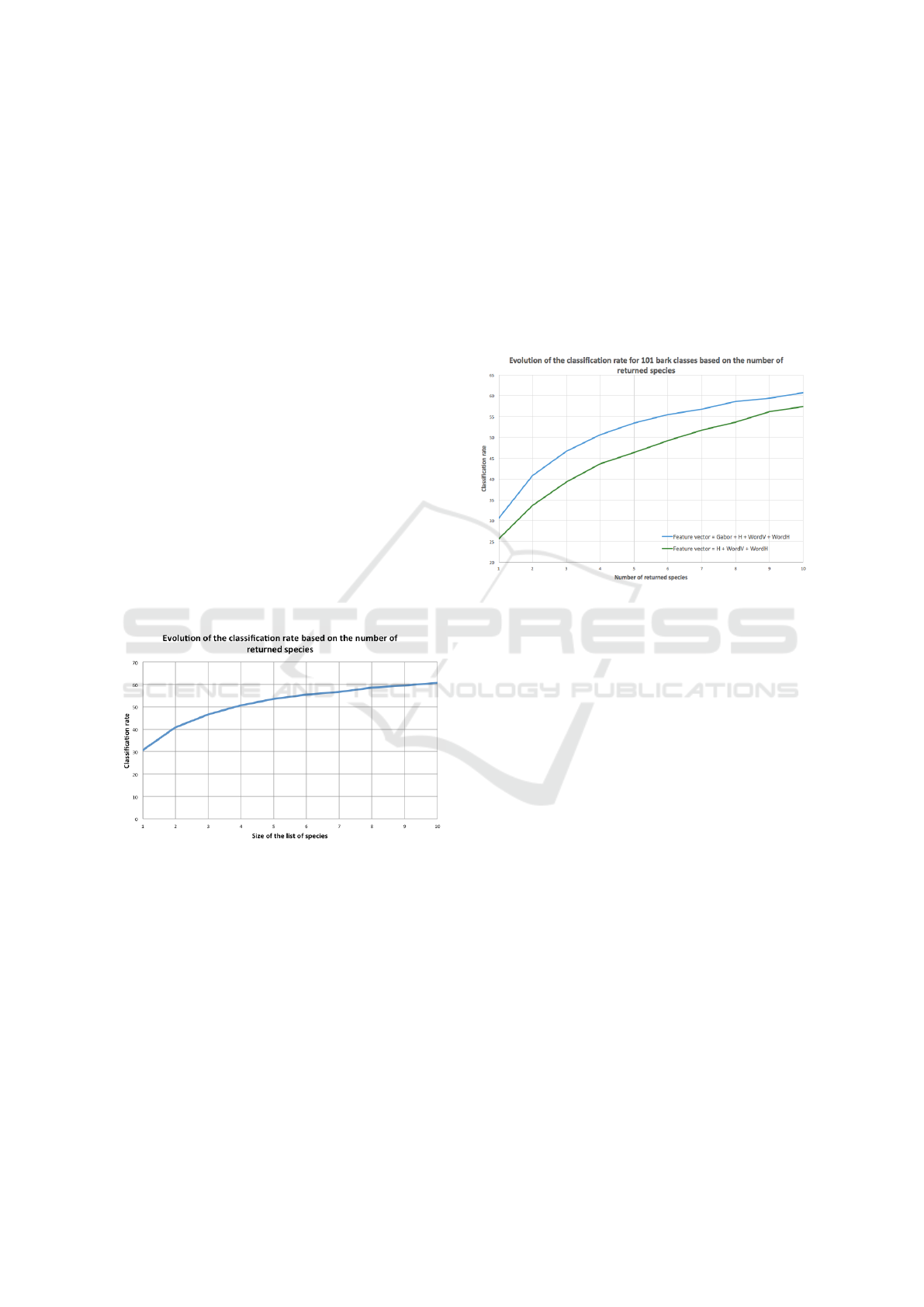
likely match the tree he photographed. The Figure 8
shows the evolution of the classification rate accord-
ing to the number of top answers returned. As said be-
fore, for a query bark image, we compute the probab-
ility of belonging to a species for each 1-vs-all binary
SVM. We order them in decreasing order the probab-
ility and check if the true species is in the top one, top
two, top three, and so on until top ten.
Figure 8: Evolution of the classification rate.
Obviously the classification rate increase with the
number of proposed species. It gains 10% when we
consider two species, and we have one chance out of
two to have the true species with a list of 4 proposi-
tions. Then progression of the recognition rate slow
down. Thus, at the end, the right species has a 60,8%
chance of being among the first ten returned species.
3.2 Discussion Concerning the Influence
of Our Bark Features
We would like to give details of the choices of our
features vectors. First, to describe the structure of the
bark, we developed W
H
and W
V
, and add information
from bark image filtered with Gabor. We can think
that the two ”vertical and horizontal words” already
capture the information given by Gabor. So we ana-
lyzed the classification results when the feature vector
is composed of the two ”words”, the hue canal and
Gabor vector, and when it is composed of only the
two ”words” and the hue canal. Results are visible
on Figure 9, which represents the classification rate
obtained if we return to the user a list of one, two or
more possible species.
Figure 9: Evolution of the classification rate with and
without the features extracted through Gabor.
As we can see, the classification rates when we
do not take into account the Gabor vector are inferior
compared to the ones obtained with the complete fea-
ture vector. Indeed, we lose on average 5% of right
classification on the classification rates depending on
the size of the list of returned species.
On the other hand, we studied the influence of the
frequency of the grid used to compute the ”vertical
word” W
V
and the ”horizontal word” W
H
. We com-
puted the classification rate while varying the number
of columns and lines of the grid respectively corres-
ponding to W
H
and W
V
. We plan to give the user of
the smart-phone application a list of five possible spe-
cies. On Figure 10, we can see the evolution of the
classification rate for the top 5 returned species.
Our results in section 3.1 are obtained with a grid
composed of 70 columns and 50 rows. We are limited
to 70 columns because the pixel widths of some of our
train bark images are too small. On Figure 10, we can
see that the best classification rate is obtained when
the number of rows of the grid is 50 and the num-
ber of columns is 70. When the number of columns
is reduced, we can see that the classification rate de-
creases. Finally, the finer the grid is, the higher the
classification rate is because the grid gets precise in-
formation.
Bark Recognition to Improve Leaf-based Classification in Didactic Tree Species Identification
439
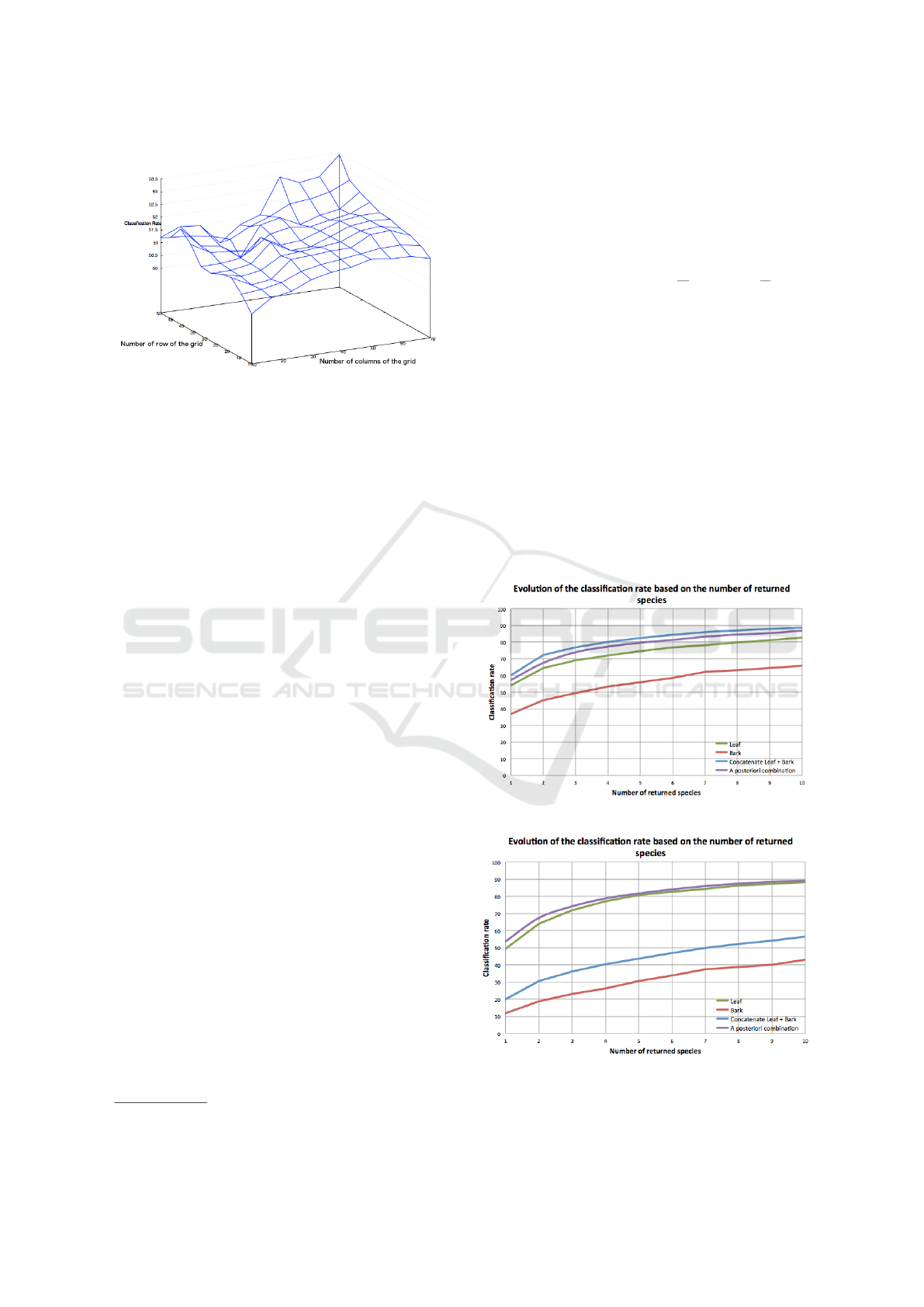
Figure 10: Influence of the frequency of the grid for the
computation of W
H
and W
V
.
3.3 Influence of Bark Processing in Tree
Leaves Classification
Most of the time, the leaves and the bark of a tree can
be observable by a person who wants to know the spe-
cies of a tree. In order to take advantage of this double
source of information, we combine the recognition of
the bark with the recognition of the leaf previously
developed and integrated to Folia
2
, the leaf identifica-
tion mobile application based on the work of (Cerutti
et al., 2013). We propose two combination methods.
First we concatenate the features of the leaf and the
features of the bark, and we use either the 1-vs-all
SVM classifier or the classifier used in the Folia ap-
plication (see (Cerutti et al., 2013)) for more inform-
ation). Secondly, we make an a posteriori combina-
tion, i.e. we do classifications independently for the
bark and for the leaf, then we combine the results of
the two classifications.
The Folia application uses a Gaussian nearest
neighbor classification in which the classifier builds a
model of each species S as a multi-dimensional Gaus-
sian distribution of its feature vectors, and estimates
the distance of a new sample represented by its fea-
ture vector F
l
to all the species models. To avoid that
the species with a high variability absorb less vari-
able classes, the distance d
GNN
(F
l
, S) is computed as
the Euclidean distance to the surface of the hyper-
ellipsoid defined by the centroid and covariance ma-
trix of the species. The result of this classification is
the species that achieves the minimal distance. The
same classification approach can be used for the bark
features F
b
and the concatenated feature vector F
b,l
.
Our a posteriori combination method uses all the
answers of the classifiers under the form of confi-
dences (between 0 and 1) associated with each spe-
2
http://liris.univ-lyon2.fr/reves/folia
cies. The SVM classifier already returns a confi-
dences value c
SV M
(F, S) and a similar value can be
computed from the Folia classifier: c
GNN
(F, S) =
e
−
d
GNN
(F,S)
2
. Then, the combination is performed
as a multiplicative fusion of the confidence values for
each species:
⊗
c (F
b
, F
l
, S) =
c(F
b
, S)
1
ω
b
c(F
l
, S)
1
ω
l
(5)
with ω
l
and ω
b
the leaf and bark weighting (6)
The resulting confidence
⊗
c is used to rank the spe-
cies and the classification answer is the species cor-
responding to the highest combined confidence.
We tested these two combination approaches on
a dataset of couples of bark and leaf images corres-
ponding the same species, for which we extracted re-
spectively the bark features presented in this article
and the leaf shape features from the folia application.
The leaf images are also from the Pl@ntView data-
base. However, some tree species have leaf image but
no bark image, and vice versa. Thereby, the number
of species we consider is reduced to 72.
Figure 11: Classification with 1-vs-All SVM classifier.
Figure 12: Classification with k-nn classifier from Folia.
On Figure 11 and Figure 12, we can see the evo-
lution of the classification rate for the different me-
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
440

thods. As we have reduced the number of classes, we
observe as expected an improvement in the bark clas-
sification rate (red curve on Figure 11). But we can
see that the classifier used in the Folia application is
really not suitable for the bark feature vector (Figure
12). For the leaf classification (green curve), the 1-
vs-all SVM classifier improves the classification rate
for the top 1 predicted species, but from the top 3 spe-
cies, the Folia classifier gives better results. We can
note that when the two feature vectors (leaf+bark) are
concatenated (blue curves), the SVM classifier (Fig-
ure 11) gives doubtlessly higher result than the Folia
classifier (Figure 12). The classifier used in Folia is in
fact not really suitable for longer vector.
Regarding the a posteriori combination (purple
curves), taking into account the bark improves the
classification rate when the leaf and the bark are re-
cognized with the Folia classifier (Figure 12). But the
performance are below the result with the concatenate
vector with SVM (Figure 11). With the classifier used
for the bark (one-vs-all SVM, Figure 11), the a poste-
riori combination show an improvement in the tree re-
cognition when we take into account the bark and not
only the leaf (purple curve). Yet it gives slightly worse
results than when we supply the SVM with a conca-
tenated feature vector of leaf and bark (blue curve).
In conclusion, we reach the best classification res-
ults when the features vectors of the bark and the leaf
are concatenated and classified with the 1-vs-all SVM
classifier.
4 CONCLUSION
In this article, we proposed an original method for
the identification of plant species to recognize trees
through the bark based on features used by botanists.
Our results achieve the same level as the state of the
art with a relatively short vector. We consider that
our method can be run with a limited computational
power and bearable by a smart-phone.
We also showed that taking into account the bark
can improve the recognition rate of trees obtained
only via the leaves. In a further work, we plan to im-
prove the combination of the bark classification and
the leaf classification by considering the results of the
confusion matrix. As a matter of fact, if we take a
look to the confusion matrix of our bark classification
on Figure 13, we can notice that some barks are more
predicted than they should. Indeed, we can clearly
see some columns where there are a lot of boxes are
green. It is the case for the classes where we have a
lot of samples and a lot of variability in the bark. In a
further work, we plan to take into account the results
in this matrix to moderate the belief in bark results
versus leaf results.
Figure 13: Confusion matrix: the greener the case, the
greater the occurrences.
Moreover, as we plan to create an educational
botanical smart-phone application, we would like to
translate the data from our features vectors as tan-
gible information for the user. For example, from the
hue histogram H, we would like to tell the user that
the bark of the tree is brown, gray, or yellowish, and
with G, W
V
and W
H
tell about the structure of the bark
(scales, strips, ridges, smooth, etc). Giving these in-
formation to the novice should help him to recognize
tree species on his own.
Furthermore, we developed W
V
and W
H
for tree
bark recognition, but we think that this concept could
be applied to other contexts where texture character-
ization is important.
Finally, in order to further improve the tree recog-
nition, we also want to extract features on the other
organs of the tree, such as the flowers, which are the
reproductive organs of the plant and therefore charac-
teristic of the species, and also the fruits, which are
characteristic of the genus.
ACKNOWLEDGMENTS
This work is part of ReVeRIES project (Reconnais-
sance de Vegetaux Recreative, Interactive et Educat-
ive sur Smartphone) supported by the French Na-
tional Agency for Research with the reference ANR-
15-CE38-004-01.
Bark Recognition to Improve Leaf-based Classification in Didactic Tree Species Identification
441

REFERENCES
Bakic, V., Mouine, S., Ouertani-Litayem, S., Verroust-
Blondet, A., Yahiaoui, I., Go
¨
eau, H., and Joly,
A. (2013). Inria’s participation at imageclef 2013
plant identification task. In CLEF (Online Working
Notes/Labs/Workshop) 2013.
Bay, H., Tuytelaars, T., and Van Gool, L. (2006). Surf:
Speeded up robust features. In Computer vision–
ECCV 2006, pages 404–417. Springer.
Canny, J. (1986). A computational approach to edge detec-
tion. Pattern Analysis and Machine Intelligence, IEEE
Transactions on, (6):679–698.
Cerutti, G., Tougne, L., Mille, J., Vacavant, A., and Coquin,
D. (2013). Understanding leaves in natural images–
a model-based approach for tree species identifica-
tion. Computer Vision and Image Understanding,
117(10):1482–1501.
Go
¨
eau, H., Bonnet, P., and Joly, A. (2015). Lifeclef plant
identification task 2015. CEUR-WS.
Go
¨
eau, H., Bonnet, P., Joly, A., Baki
´
c, V., Barbe, J.,
Yahiaoui, I., Selmi, S., Carr
´
e, J., Barth
´
el
´
emy, D.,
Boujemaa, N., et al. (2013a). Pl@ntnet mobile app.
In Proceedings of the 21st ACM international confer-
ence on Multimedia, pages 423–424. ACM.
Go
¨
eau, H., Bonnet, P., Joly, A., Bakic, V., Barth
´
el
´
emy, D.,
Boujemaa, N., and Molino, J.-F. (2013b). The image-
clef 2013 plant identification task. In CLEF.
Go
¨
eau, H., Bonnet, P., Joly, A., Boujemaa, N., Barth
´
el
´
emy,
D., Molino, J.-F., Birnbaum, P., Mouysset, E., and Pi-
card, M. (2011). The imageclef 2011 plant images
classi cation task. In ImageCLEF 2011, pages 0–0.
Go
¨
eau, H., Joly, A., Bonnet, P., Selmi, S., Molino, J.-F.,
Barth
´
el
´
emy, D., and Boujemaa, N. (2014). Lifeclef
plant identification task 2014. In CLEF2014 Work-
ing Notes. Working Notes for CLEF 2014 Conference,
Sheffield, UK, September 15-18, 2014, pages 598–
615. CEUR-WS.
Huang, Z.-K., Huang, D.-S., Du, J.-X., Quan, Z.-H., and
Guo, S.-B. (2006). Bark classification based on gabor
filter features using rbpnn neural network. In Neural
Information Processing, pages 80–87. Springer.
Kumar, N., Belhumeur, P. N., Biswas, A., Jacobs, D. W.,
Kress, W. J., Lopez, I. C., and Soares, J. V. (2012).
Leafsnap: A computer vision system for automatic
plant species identification. In Computer Vision–
ECCV 2012, pages 502–516. Springer.
Lowe, D. G. (1999). Object recognition from local scale-
invariant features. In Computer vision, 1999. The pro-
ceedings of the seventh IEEE international conference
on, volume 2, pages 1150–1157. Ieee.
Nakayama, H. (2013). Nlab-utokyo at imageclef 2013 plant
identification task. In Working notes of CLEF 2013
conference.
Ojala, T., Pietik
¨
ainen, M., and Harwood, D. (1996). A com-
parative study of texture measures with classification
based on featured distributions. Pattern recognition,
29(1):51–59.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S.,
Anguelov, D., Erhan, D., Vanhoucke, V., and Ra-
binovich, A. (2015). Going deeper with convolutions.
In Proceedings of the IEEE Conference on Computer
Vision and Pattern Recognition, pages 1–9.
Vapnik, V. N. (1999). An overview of statistical learn-
ing theory. Neural Networks, IEEE Transactions on,
10(5):988–999.
Wojtech, M. (2011). Bark: A Field Guide to Trees of the
Northeast. University Press of New England.
VISAPP 2017 - International Conference on Computer Vision Theory and Applications
442
