
An Accuracy Comparison of the Joint and Sequential Approaches for
End-to-End Related Named Entities Extraction in the Texts of
Russian-Language Reviews Based on Neural Networks
Sboev Alexander
1,2 a
, Roman Rybka
1,3 b
, Aleksandr Naumov
1 c
,
Anton Selivanov
1 d
, Artem Gryaznov
1 e
and Ivan Moloshnikov
1
1
National Research Centre “Kurchatov Institute”, Academic Kurchatov sq., Moscow, Russian Federation
2
National Research Nuclear University “MEPhI”, Kashirsk. hw., Moscow, Russian Federation
3
Russian Technological University “MIREA”, Vernadsky av., Moscow, Russian Federation
Keywords:
Named Entity Recognition, Relation Extraction, Joint Model, Russian Drug Review Corpus, Deep Learning,
Language Models, Natural Language Processing, Pharmacovigilance.
Abstract:
Solving a problem of relations recognition among significant pharmacological entities is one of the important
stage of complex automatic analysis of drug reviews for purposes of pharmacovigilance, marketing, social
situation analysis, healthcare, and others. The closest statement of the problem to practical cases is an end-to-
end model to extract related entities from the scratch, with realization of two stages: recognition of significant
entities (NER) and extraction of relation between them (RE). To our knowledge, this problem has not been
solved for Russian drug review texts of every day lexis. So, there is no evaluation of the accuracy of its solution.
A creation of the Russian Drug Review Corpus RDRS allowed to obtain such an evaluation presented in this
work. We use two models for this purpose: the first is of joint NER and RE extraction, the second is of the
step by step calculations, initially of NER and then RE. The difference in results, obtained on the basis of
the above two ways, was analyzed. Both approaches demonstrated the close average accuracies of end-to-end
solution, establishing an accuracy level of the problem in view about 51% f1 for the set of related entities:
ADR-Drugname, Drugname- Diseasename, Drugname- SourceInfoDrug, Diseasename- Indication.
1 INTRODUCTION
Currently, the exchange of the information among
users in the Internet environment through specialized
platforms and social networks is widespread. Collect-
ing and analyzing this information provide the basis
for conducting large-scale user experience studies, re-
garding user reactions to the use of medicines. The
importance of this task is determined by the need to
monitor the consequences of the use of drugs in the
post-clinical period, both by specialized state bodies
(pharmacovigilance) and by pharmaceutical manufac-
turing companies. The extraction of this information
from the reviews of Internet users is based on the
identification of related named entities in natural lan-
a
https://orcid.org/0000-0002-6921-4133
b
https://orcid.org/0000-0002-5595-6398
c
https://orcid.org/0000-0002-4114-4460
d
https://orcid.org/0000-0001-5075-7229
e
https://orcid.org/0000-0003-0449-4549
guage texts, which are characterized by the presence
of typos, omissions of punctuation marks, the use of
informal language, slang, jargon, the lack of standard-
ized terminology, different cases of using the same or
several drugs in one review. The recent researches
show that the greatest efficiency in a neural net end-
to-end solution of the problem NER and RE of natu-
ral language texts is achieved by two mainstream ap-
proaches: sequential one (Zhang et al., 2019; Sahu
and Anand, 2018; Quan et al., 2016) and joint solu-
tion (Li et al., 2017; Henry et al., 2019; Eberts and
Ulges, 2020; Fang et al., 2021). A joint approach, on
the one hand, allows getting a synergistic effect from
the joint learning both tasks simultaneously, on the
other hand, it requires finer tuning to solve both prob-
lems effectively. Thus, a comparative analysis of the
sequential and joint approaches to solving the prob-
lem of NER and RE is demanded to select a preferable
approach for further evaluation of the State-of-the-Art
(SoTA) level of problem in view for Russian drug re-
348
Alexander, S., Rybka, R., Naumov, A., Selivanov, A., Gryaznov, A. and Moloshnikov, I.
An Accuracy Comparison of the Joint and Sequential Approaches for End-to-End Related Named Entities Extraction in the Texts of Russian-Language Reviews Based on Neural Networks.
DOI: 10.5220/0011926900003612
In Proceedings of the 3rd International Symposium on Automation, Information and Computing (ISAIC 2022), pages 348-353
ISBN: 978-989-758-622-4; ISSN: 2975-9463
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

view texts.
The main contributions of this work are:
• comparative evaluations of accuracies of the se-
quential and joint approaches on the base of Rus-
sian Drug Review Corpus (RDRS);
• establishing the State-of-the-Art level accuracy of
solving the problem NER and RE for the Russian
natural language text of drug reviews.
Further, in the Section 2.1 the description of
the corpus of Russian-language reviews used is pre-
sented. In the Section 2.2 both approaches are de-
scribed, and the language models in their composi-
tion. The Section 3 provides a description of the
experiments and the evaluation procedure. The Sec-
tions 4 and 5 describes the results of experiments and
conclusions on the work as a whole.
2 MATERIALS AND METHODS
2.1 Data
The study is based on the Russian Drug Review Cor-
pus (RDRS) (Sboev et al., 2022). The corpus con-
tains texts of user reviews about medicines from the
site Otzovik.ru. Each review is annotated by ex-
perts with pharmaceutical education under the cross-
checking procedure (see original work). Annotations
include several types of markup: 1) named entities,
2) different cases of drug use (each individual case is
hereinafter referred to as “context”). More than 20
types of named entities are distinguished in the cor-
pus, which can be attributed to the three categories:
• Medication – entities of this category describe
medication and it’s attributes: drugname, class,
form, dosage, way of use etc;
• Disease – entities that describe symptoms, a dis-
ease and dynamics of condition;
• Adverse Drug Reaction – entities that describe the
adverse reactions to drug use mentioned by the au-
thors.
Each named entity was referred by annotators to one
of the contexts that differed: description of different
cases of using one drug, comparison of cases of using
different drugs, as well as different symptoms of dis-
eases. An example of a review with annotations for
named entities and contexts is shown in the Figure 1.
The main corpus of RDRS consists of 2800 testimo-
nial texts and their annotations, which were divided
into 5 subsets of training and test cases (folds). In this
paper, we have chosen 4 types of related entities that
are of greater practical interest:
• ADR–Drugname – adverse effect of the particular
medication;
• Drugname–SourceInfodrug – source of the infor-
mation about medication (e.g. “my brother gave
me advice”, “apothecary mentioned”);
• Drugname–Diseasename – a link between the
disease and medication that user administrated
against it;
• Diseasename–Indication -— symptoms of the
particular disease (e.g., “red rash”, “high temper-
ature”).
Number of entities, relations and other statistic are
shown in Tables 1, 2 and 3.
2.2 Methods and Approaches
2.2.1 Sequential Approach
A sequential solution of the end-to-end problem of
RE extraction is the consistent application of two
models (Figure 2): the first extracts entities and the
second detects relations. Entity extraction is based
on a multivalued classification of tokens using BIO
markup, which we successfully tested in our previous
work (Sboev et al., 2022). Each token is marked as
“B-classname” - the initial token of the “classname”
class entity, “I-classname” - the token belonging to
the “classname” class entity, “O” - the token not be-
longing to named entities. The model itself consists
of a pre-trained language model with transformer ar-
chitecture, and classification layers. The output activ-
ities of the last layer of the language model are fed
into several linear layers, each of which corresponds
to a separate class of named entities and has 3 output
neurons for labels B, I, and O with a softmax activa-
tion function. The task of entity extraction solves as
a pair of entities classification. To classify the pairs,
a pre-trained language model of transformer architec-
ture with a classification layer was used. The model
input is the text of the review and selected entities in
a special format.
1. Concatenate the pair of considered entities
through special [ESEP] token;
2. Concatenated obtained sequence with the text that
contains entities, special token [TXTSEP] is used
to sepate entities and text;
3. Add special token [CLS] to the sequence, the
model is trained in such way that vector of this
token aggregates information abouth whole se-
quence and is used to classify an entity pair;
An Accuracy Comparison of the Joint and Sequential Approaches for End-to-End Related Named Entities Extraction in the Texts of
Russian-Language Reviews Based on Neural Networks
349
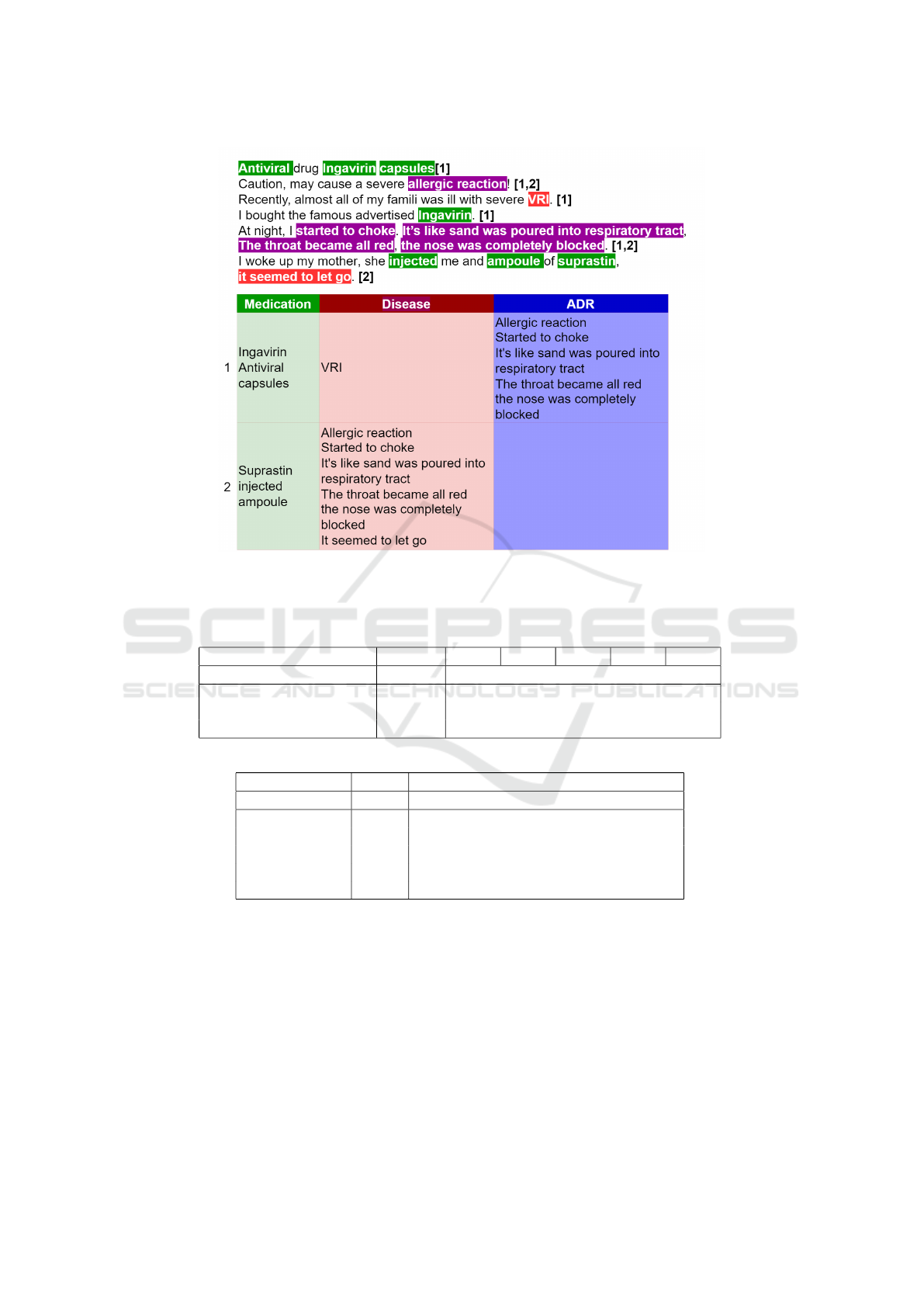
Figure 1: Example of annotation with entities separated into 2 contexts. Number after each sentence shows to which context
entities in the sentence were assigned. Phrases highlighted in green are mentions of medication attributes, red color indicates
disease, symptoms and dynamics, blue color is used for ADR mentions, phrases highlighted with purple color are annotated
both as Disease and ADR but included in different contexts.
Table 1: Number of texts and their length statistic for RDRS corpus.
Value all folds fold 1 fold 2 fold 3 fold 4 fold 5
text number 2798 559 559 560 560 560
avg text length (in words) 157 157 159 156 156 156
min text length 42 48 49 46 47 42
max text length 276 256 248 246 240 276
Table 2: Number of entities of different types in the RDRS corpus.
Entity number total fold 1 fold 2 fold 3 fold 4 fold 5
Total 21740 4420 4328 4298 4365 4329
ADR 1809 380 390 329 382 328
Drugname 8467 1695 1664 1736 1686 1686
SourceInfodrug 2595 530 523 508 519 515
Diseasename 4026 801 769 760 842 854
Indication 4843 1014 982 965 936 946
2.2.2 Joint Approach
The scheme of model of joint approach shown on the
Figure 3. After tokenization, the input text is added
by special context token t
c
to code the whole text.
Next, the resulting sequence of tokens and a special
token are vectorized using the language model. Un-
like the NER model from the sequential approach, in
this model, the definition of named entities is based
on the classification of spans. S
M
spans are se-
quences of consecutive tokens. The spans can in-
tersect and have different sizes, but no more than
the specified maximum. The vector representation
for each span is formed by concatenating the vec-
tor representation of the span length and the vector
obtained from the vectors of tokens included in the
span: s
i j
= maxpool(t
i
, t
i+1
, ...t
j
)∗Embedding( j − i).
The concatenated vectors of the span s
i j
and the vec-
tor of context token t
c
are fed into a fully connected
layer, in which the number of output neurons is equal
ISAIC 2022 - International Symposium on Automation, Information and Computing
350
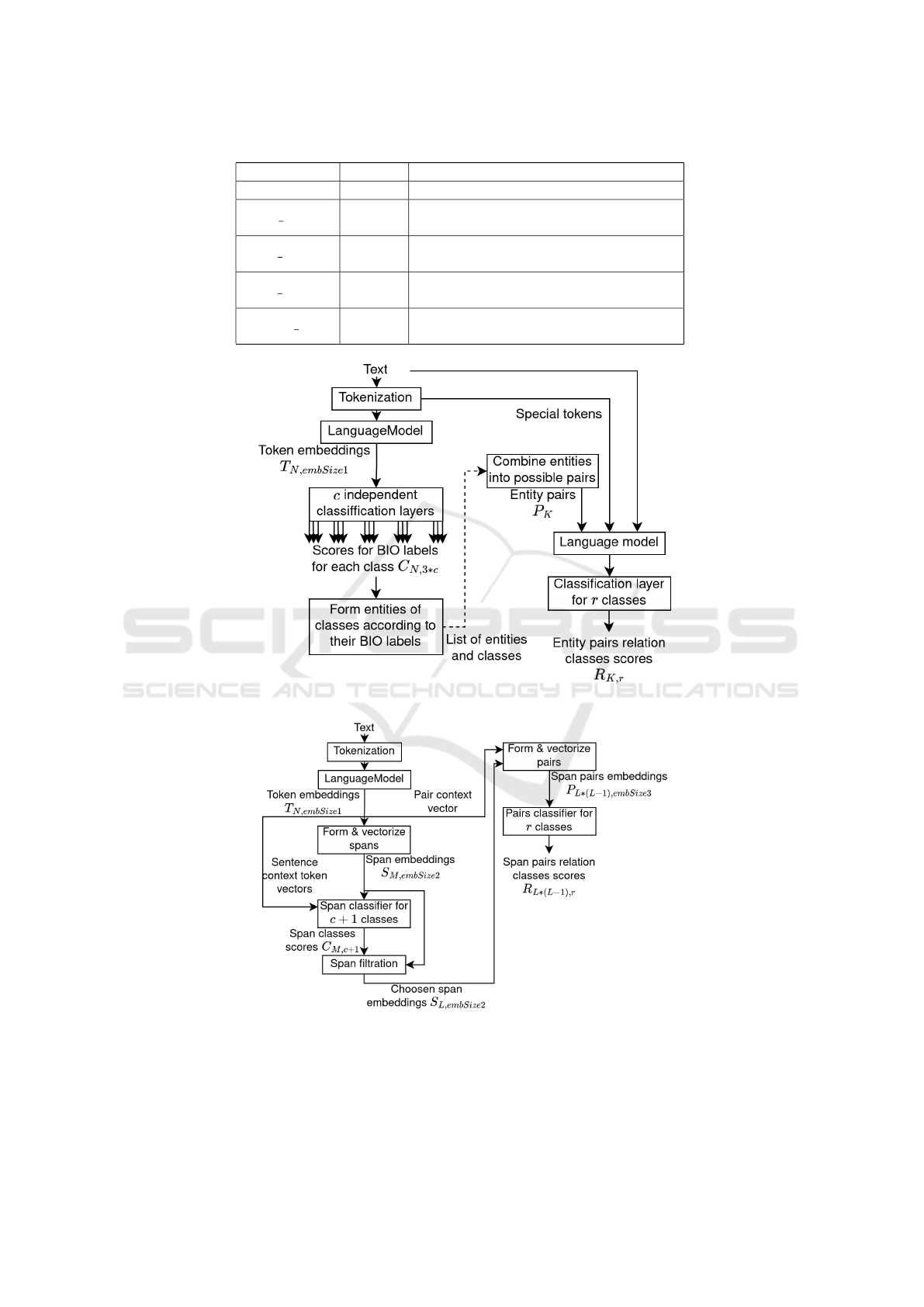
Table 3: Number of relations of different types in the RDRS corpus.
Relation nb all folds fold 1 fold 2 fold 3 fold 4 fold 5
Total 34597 6952 6501 6855 7257 7032
ADR Drug
4274
(910)
844
(166)
832
(168)
812
(210)
1004
(212)
782
(154)
Drug Disease
11135
(2108)
2168
(383)
2107
(361)
2138
(429)
2311
(541)
2411
(394)
Drug Info
7056
(1279)
1481
(297)
1404
(229)
1381
(316)
1382
(225)
1408
(212)
Disease Ind
7093
(742)
1469
(144)
1223
(177)
1443
(126)
1446
(136)
1512
(159)
Figure 2: Scheme of sequential approach. Dotted line illustrates dataflow between 2 separate models.
Figure 3: Scheme of joint approach.
to the number of entity classes with the addition of
the notEntity class for spans that are not named en-
tities. The outputs of a fully connected layer are
normalized by the softmax function. All spans of
the notEntity class are not used in further calcula-
tions. Remaining spans participate in the pairing and
An Accuracy Comparison of the Joint and Sequential Approaches for End-to-End Related Named Entities Extraction in the Texts of
Russian-Language Reviews Based on Neural Networks
351

a classification procedure is performed. All possible
pairs are compiled from the list of selected spans and
their vector representation is formed by concatenat-
ing: a) the span vectors in the pair and b) the local
context vector of the pair p, which is calculated as
follows: p = s
i j
maxpool(t
j+1
, ..., t
i
′
−1
) ∗ s
i
′
j
′
, where
t
j+1
, ..., t
i
′
−1
are the tokens between the last and the
first token of the named entities of the pair in ques-
tion. Paired vectors are fed into a fully connected
layer with output neurons with sigmoid activation cor-
responding to connection classes. All pairs for which
the output activity of at least one neuron was above
the threshold are considered to be present in the text
and have a class corresponding to the neuron. Pairs
for which none of the actions exceeded the threshold
are considered unrelated.
2.2.3 Language Models
XLM-RoBERTa-large (Liu et al., 2019) (further
XLMR) - a language model with a transformer ar-
chitecture, which, like BERT (Devlin et al., 2018),
was trained on the problem of predicting masked to-
kens and the task of predicting the next sentence. But
RoBERTa had significantly more training data and
modified training hyperparameters. The RoBERTa
training set had 160 GB of texts in different lan-
guages, including corpora: BookCorpus (Zhu et al.,
2015), English Wikipedia
1
, CC-News (Hamborg
et al., 2017), OpenWebText
2
, Stories (Trinh and Le,
2018). XLM-RoBERTa-large version contains 550M
weights.
XLM-RoBERTa-sag (further XLMR sag) is a ver-
sion of the RoBERTa-large model adapted to pharma-
ceutical product reviews (Sboev et al., 2022). To this
purpose, it was further trained on the corpus
3
contain-
ing 2 sets of texts: the first contained 250,000 drug
reviews and was collected from the irecommend.ru
website, the second set was taken from not annotated
part of RuDReC.
3 EXPERIMENTS
Experiments to compare both approaches are per-
formed on the RDRS corpus datasets on base of cross-
validation check on 5 folds. As a result, both named
entities and relationships between them are automat-
ically extracted, so the accuracy assessment is car-
ried out immediately for both solutions as part of
a common task. A well-defined named entity is a
1
https://en.wikipedia.org/wiki/
2
http://Skylion007.github.io/OpenWebTextCorpus
3
https://huggingface.co/sagteam/xlmroberta-large-sag
phrase whose boundaries and class match the refer-
ence markup in the source corpus. A correct selection
of related named entities is pairs of entities whose
boundaries and classes coincide with the reference
markup, and the presence of relationships between
them, is correctly determined.
4 DISCUSSION
Obtained results of 51-52% f1-macro (see Fig. 4 and
Table 4) for RE show an agreement in the accuracies
of both approaches, taking into the account the devia-
tion of 1% from the average on five-fold cross valida-
tion for all runs. At the same time, the accuracy level
of entity identifications in composition of sequential
approach is lower in relation to joint one (see Table 5).
But, as concerns relation identifications, the situation
is vice versa, that in general, gives close results.
5 CONCLUSION
This paper sets first the current level of accuracy of
end-to-end solving the related entities’ extraction
task for Russian review texts using the RDRS cor-
pus by two approaches: joint and sequential. The
established accuracy level of the problem in view
about 51% f1 for the set of related entities: ADR-
Drugname, Drugname-Diseasename, Drugname-
SourceInfoDrug, Diseasename-Indication. This value
may be a reference point for a further moderniza-
tion of end-to-end models for Russian and will be
considered in our future works. The results obtained
expand the set of solutions for analyzing the texts of
Internet user reviews about pharmaceutical products
and can become the basis for building a system for
automatically monitoring adverse reactions that occur
when taking drugs, and describing cases of their use.
ACKNOWLEDGEMENTS
The study was supported by a grant from the Rus-
sian Science Foundation (project no. 20-11-20246).
This work has been carried out using computing re-
sources of the federal collective usage center Com-
plex for Simulation and Data Processing for Mega-
science Facilities at NRC “Kurchatov Institute”,
http://ckp.nrcki.ru/
ISAIC 2022 - International Symposium on Automation, Information and Computing
352
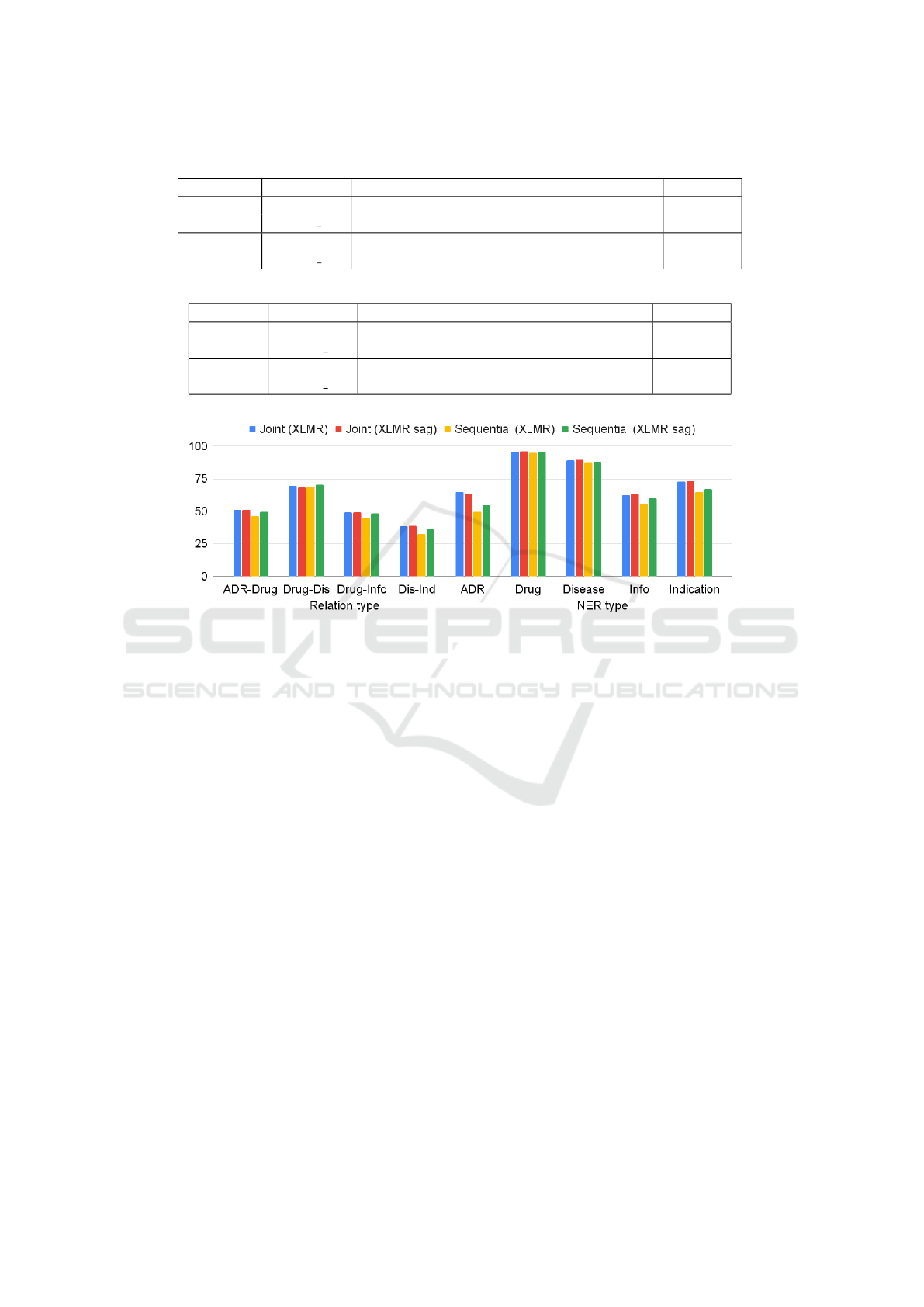
Table 4: Evaluation scores for named relation extraction task. ADR - Adverse Drug Reaction, Drug - Drugname, Dis -
Diseasename, Info - SourceInfoDrug, Ind - Indication
Approach Model ADR-Drug Drug-Dis Drug-Info Dis-Ind f1-macro
Joint XLMR 51.2 69.4 49.2 38.6 52.1
Joint XLMR sag 51.1 68.3 49 38.9 51.8
Sequential XLMR 46.1 69.2 45.1 32.2 48.1
Sequential XLMR sag 49.4 70.4 48.3 36.7 51.2
Table 5: Evaluation scores for named entity recognition task.
Approach Model ADR Drug Disease Info Indication f1-macro
Joint XLMR 64.8 95.7 89.4 62.5 72.9 77.1
Joint XLMR sag 63.8 96.0 89.7 63.3 73.2 77.2
Cascade XLMR 49.6 95.1 87.7 55.6 64.7 70.5
Cascade XLMR sag 54.7 95.3 88.3 60.0 67.2 73.1
Figure 4: Evaluation scores for different language models.
REFERENCES
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Eberts, M. and Ulges, A. (2020). Span-based joint entity
and relation extraction with transformer pre-training.
In ECAI 2020, pages 2006–2013. IOS Press.
Fang, X., Song, Y., and Maeda, A. (2021). Joint extrac-
tion of clinical entities and relations using multi-head
selection method. In 2021 International Conference
on Asian Language Processing (IALP), pages 99–104.
IEEE.
Hamborg, F., Meuschke, N., Breitinger, C., and Gipp, B.
(2017). news-please: A generic news crawler and ex-
tractor. In Proceedings of the 15th International Sym-
posium of Information Science, pages 218–223.
Henry, S., Buchan, K., Filannino, M., Stubbs, A., and
Uzuner, O. (2019). 2018 n2c2 shared task on adverse
drug events and medication extraction in electronic
health records. Journal of the American Medical In-
formatics Association, 27(1):3–12.
Li, F., Zhang, M., Fu, G., and Ji, D. (2017). A neural joint
model for entity and relation extraction from biomed-
ical text. BMC bioinformatics, 18(1):1–11.
Liu, Y., Ott, M., Goyal, N., Du, J., Joshi, M., Chen, D.,
Levy, O., Lewis, M., Zettlemoyer, L., and Stoyanov,
V. (2019). Roberta: A robustly optimized bert pre-
training approach. arXiv preprint arXiv:1907.11692.
Quan, C., Hua, L., Sun, X., and Bai, W. (2016). Multichan-
nel convolutional neural network for biological rela-
tion extraction. BioMed research international, 2016.
Sahu, S. K. and Anand, A. (2018). Drug-drug interac-
tion extraction from biomedical texts using long short-
term memory network. Journal of biomedical infor-
matics, 86:15–24.
Sboev, A., Sboeva, S., Moloshnikov, I., Gryaznov, A., Ry-
bka, R., Naumov, A., Selivanov, A., Rylkov, G., and
Ilyin, V. (2022). Analysis of the full-size russian cor-
pus of internet drug reviews with complex ner label-
ing using deep learning neural networks and language
models. Applied Sciences, 12(1):491:1–34.
Trinh, T. H. and Le, Q. V. (2018). A simple
method for commonsense reasoning. arXiv preprint
arXiv:1806.02847.
Zhang, T., Lin, H., Ren, Y., Yang, L., Xu, B., Yang, Z.,
Wang, J., and Zhang, Y. (2019). Adverse drug re-
action detection via a multihop self-attention mech-
anism. BMC bioinformatics, 20(1):1–11.
Zhu, Y., Kiros, R., Zemel, R., Salakhutdinov, R., Urtasun,
R., Torralba, A., and Fidler, S. (2015). Aligning books
and movies: Towards story-like visual explanations by
watching movies and reading books. In The IEEE In-
ternational Conference on Computer Vision (ICCV).
An Accuracy Comparison of the Joint and Sequential Approaches for End-to-End Related Named Entities Extraction in the Texts of
Russian-Language Reviews Based on Neural Networks
353
