
Adaptive Adversarial Samples Based Active Learning for Medical Image
Classification
Siteng Ma
1
, Yu An
1
, Jing Wang
2
, Aonghus Lawlor
1
and Ruihai Dong
1
1
The Insight Centre for Data Analytics, School of Computer Science, University College Dublin, Dublin, Ireland
2
College of Computer Science, North China Institute of Aerospace Engineering, Langfang, China
Keywords:
Deep Learning, Active Learning, Adversarial Attack, Counterfactual Sample, Medical Image Classification.
Abstract:
Active learning (AL) is a subset of machine learning, which attempts to minimize the number of required
training labels while maximizing the performance of the model. Most current research directions regarding
AL focus on the improvement of query strategies. However, efficiently utilizing data may lead to more perfor-
mance improvements than are thought to be achievable by changing the selection strategy. Thus, we present
an adaptive adversarial sample-based approach to query unlabeled samples close to the decision boundary
through the adversarial attack. Notably, based on that, we investigate the importance of using existing data
effectively in AL by integrating generated adversarial samples according to consistency regularization and
leveraging large numbers of unlabeled images via pseudo-labeling with the oracle-annotated instances during
training. In addition, we explore an adaptive way to request labels dynamically as the model changes state.
The experimental results verify our framework’s effectiveness with a significant improvement over various
state-of-the-art methods for multiple medical applications. Our method achieves 3% above the supervised
learning accuracy on the Messidor Dataset (the task of Diabetic Retinopathy detection) using only 34% of the
whole dataset. We also conducted an extensive study on a histological Breast Cancer Diagnosis Dataset. Our
code is available at https://github.com/HelenMa9998/adversarial active learning.
1 INTRODUCTION
Deep learning (DL) techniques have achieved great
success in medical image diagnosis (Litjens et al.,
2017). However, these DL-based solutions require a
large amount of labeled data to train. Labeling data is
expert-oriented, time-consuming, and expensive, es-
pecially in the medical field, which has impeded the
development of DL in different medical image diag-
nosing tasks. Fortunately, Active learning (AL) can
mitigate this impediment by incrementally selecting
informative samples for manual annotation, resulting
in high performance with less labeling effort.
AL methods generally focus on designing query
strategies to obtain more valuable samples. The most
popular ones are designed based on the uncertainty of
model predictions (Joshi et al., 2009; Houlsby et al.,
2011). In addition, promoting the diversity of cho-
sen instances is another crucial approach (Sener and
Savarese, 2017; Gal et al., 2017), and recent works
are exploring the hybrid method to combine the cri-
terion of uncertainty and diversity (Ash et al., 2019;
Zhdanov, 2019).
With the popularity of generative networks like
Generative adversarial networks (GANs) or Varia-
tional auto-encoders (VAEs), attention has been paid
to adversarial samples. Several researchers have ex-
plored generating data with higher uncertainty to la-
bel or help the AL process (Zhu and Bento, 2017;
Tran et al., 2019; Sinha et al., 2019), but generating
plausible images remains a difficult problem and also
with high cost, especially in the medical domain. In
contrast, Ducoffe and Precioso proposed the Deep-
Fool Active Learning method (DFAL) based on an
adversarial attack by gradually adding noise to data
until being misclassified by the model (Ducoffe and
Precioso, 2018). In other words, they selected unla-
beled instances with the lowest adversarial perturba-
tions (i.e., samples closer to the decision boundary).
This approach proved effective in MNIST, the Shoe-
Bag, and the Quick-Draw datasets. However, studies
applying them to medical diagnosis are still lacking.
In addition, medical images were found to be
more vulnerable to adversarial attack compared to
natural images in paper (Ma et al., 2021), which indi-
cates that the adversarial samples (i.e., the generated
images by adding noise during the adversarial attacks)
Ma, S., An, Y., Wang, J., Lawlor, A. and Dong, R.
Adaptive Adversarial Samples Based Active Learning for Medical Image Classification.
DOI: 10.5220/0011622100003411
In Proceedings of the 12th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2023), pages 751-758
ISBN: 978-989-758-626-2; ISSN: 2184-4313
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
751

might be meaningful for the training process. There-
fore, when exploring adversarial attack-based meth-
ods in the medical field, we add unlabeled instances
and their free counterfactual samples with the same
labels to the training dataset as a data expansion skill.
Furthermore, in contrast to selected instances closer
to the decision boundary, examples far away from it
are of high confidence. Intuitively, we can harness
those unlabeled samples by pseudo labeling instead of
manual annotation to further decrease label cost. Ad-
ditionally, since the model is inconsistent throughout
the process, we propose an adaptive AL correspond-
ing to the model’s state.
Overall, we extend DFAL to reduce the annotation
effort further and improve the model performance in
the medical domain through different data utilization
skills. We committed to fully using the adversarial at-
tack principle during the process so that the model
achieves better results with as little labeled data as
possible. Our approach is validated by conducting
experiments on two medical image diagnosis tasks
and modalities: diabetic retinopathy detection from
retinal fundus images and breast cancer grading from
histopathological images. Fig. 1 shows the fundamen-
tal idea of our method, which takes the Messidor (De-
cenci
`
ere et al., 2014), a Diabetic Retinopathy detec-
tion dataset, as an example.
The main contributions in this study are therefore:
• Novelty: to the best of our knowledge, we are
the first to introduce the adversarial attack method
with AL for medical image analysis. Counterfac-
tual augmentation, pseudo labeling, and an adap-
tive AL strategy are proposed on top of this base.
• Efficiency: we achieve superior results with fewer
labeled examples than competing benchmarks and
outperform the fully supervised learning baseline.
• Robustness: our method shows consistent supe-
rior performance on both binary and multi-class
classification problems.
2 RELATED WORK
2.1 Deep Active Learning
Deep active learning (DAL), the combination of DL
and AL, can effectively solve the problem of lim-
ited labeled data. Every DAL scenario involves de-
termining the information contained in unlabeled in-
stances, defined as query strategy. There are many
proposed ways of formulating query strategies in the
literature. The most common method is uncertainty
sampling that takes confident the model prediction as
standard (Settles, 2009). One specific sample is infor-
mation entropy that unlabeled data above an thresh-
old are selected for annotation (Joshi et al., 2009).
In 2011, paper (Houlsby et al., 2011) introduced the
use of Bayesian convolutional neural networks for
AL (BALD) which calculated the difference between
the entropy of the average prediction and the average
entropy of stochastic predictions. In the same pa-
per, Monte Carlo (MC) dropout is performed to ob-
tain different class posterior probabilities in parame-
ter sets drawn from dropout distribution. However,
uncertainty-based methods are likely to ignore the re-
lationship between samples. Therefore, as another
important criterion, diversity has come out, requir-
ing annotation according to data representation: the
data that show their high diversity compared to the la-
beled can be more helpful for model training. The
Core-set technique is proposed as an effective rep-
resentation learning method to select samples (Sener
and Savarese, 2017). Then, to combine the strengths
of uncertainty and diversity methods, hybrid query
strategies (Ash et al., 2019; Zhdanov, 2019; Smailagic
et al., 2018; Smailagic et al., 2020) aim to achieve
large uncertainty and small redundancy of selected
samples. Among them, Smailagic (2018)’ technique
is applied in medical images, combining entropy and
distance between feature descriptors, and based on
this, they further explores a more effective training
method in 2020. Both of them will be compared with
our approach in section 4.3.
All these papers mentioned above are committed
to using a strategy to select the most representative
samples while ignoring the contribution of data uti-
lization to AL, either labeled or unlabeled data, caus-
ing a waste of labels. We argue that data utilization
skills can be the key to addressing the issue of the
number of AL queried samples being insufficient to
support the update of the DL models and therefore
boost the AL process. Consequently, we proposed an
adaptive way based on the adversarial attack to ex-
pand the training dataset with generated adversarial
samples and pseudo-labeled data.
2.2 Adversarial Attacks
Szegedy et al. first composed the idea of adversarial
examples, demonstrating the existence of small per-
turbations to the images (Szegedy et al., 2013). Such
perturbed samples could fool DL models into misclas-
sification but appear similar to the clean images from
a human’s perspective. More formally, given a pre-
trained network h and an original image x with label
y target, an attacking method is to maximize the clas-
sification error of the h that the prediction becomes
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
752
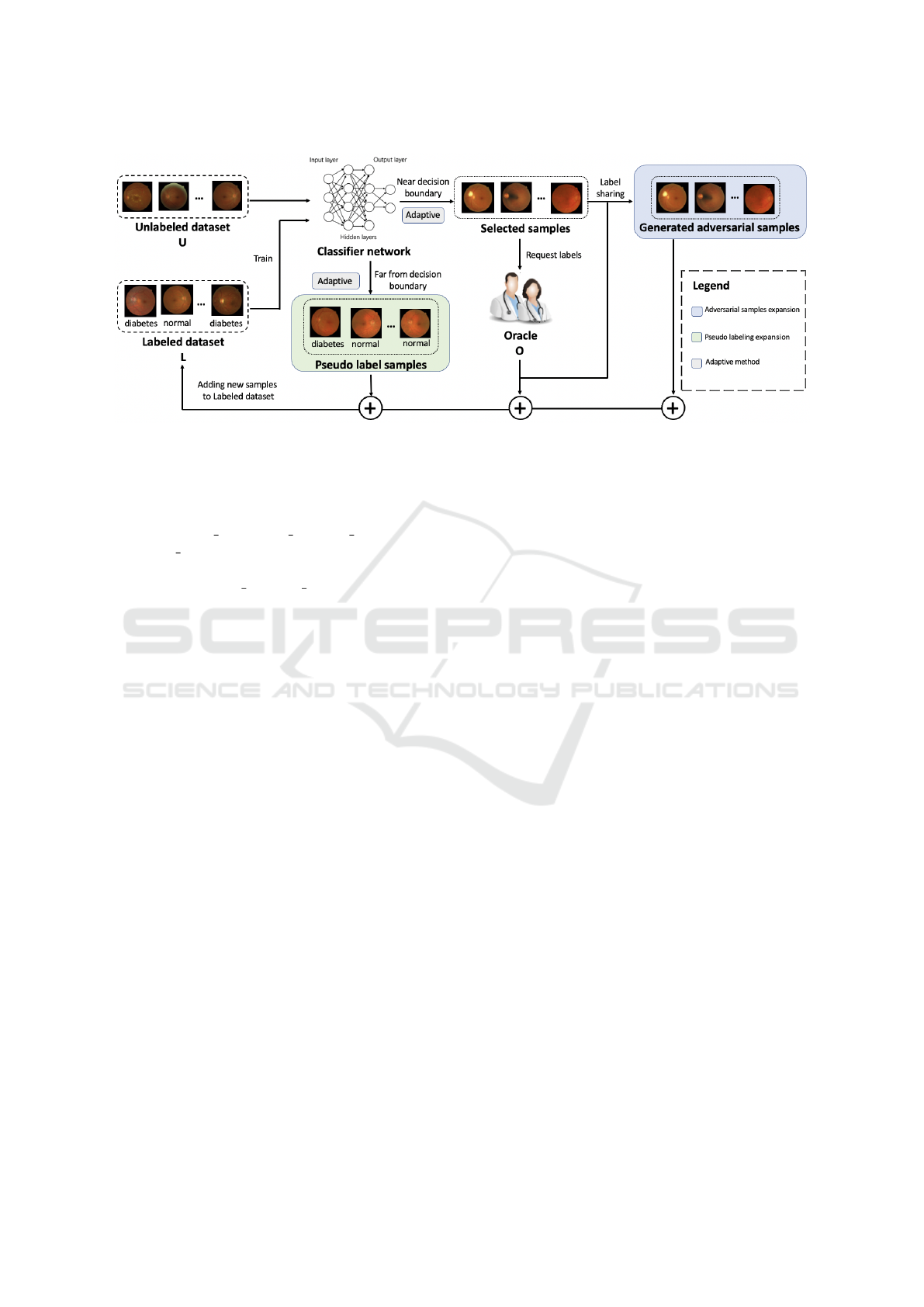
Figure 1: Overview of Adaptive Adversarial sample-based AL. It consists of four modules: (a) AL process based on an
adversarial attack by selecting the samples close to the decision boundary for labeling and adding Labeled dataset L. (b)
Adversarial samples expansion to add generated samples to L according to consistency regularization. (c) Pseudo-labeling
expansion. In contrast to the process of (a), samples far from the decision boundary are selected and added to L. (d) Adaptive
method dynamically selects samples in (a) and (c) depending on the different states of the model throughout the AL process.
different from y target (h(x adv) 6= y target), whilst
keeping x adv within a small difference ρ compared
to the original sample x, due to its consistency for hu-
man perception (h(x adv) = y target).
Furthermore, in the medical domain, recent work
(Ma et al., 2021) concludes that compared with nat-
ural image models, medical deep neural models are
more vulnerable to adversarial attack. Work (Paschali
et al., 2018) utilizes Dense Adversarial Generation to
craft adversarial examples, showing that classification
accuracy drops from 87% on original medical images
to almost 0% on adversarial examples in skin lesion
classification and whole brain segmentation. Authors
(Finlayson et al., 2019) have confirmed that diagnosis
results can be arbitrarily manipulated by an adversar-
ial attack from experiments across Fundoscopy, Chest
X-Ray, and Dermoscopy. Some papers (Ren et al.,
2019; Pervin et al., 2021) take advantage of the adver-
sarial attack for augmentation to eliminate overfitting
and improve model’s performance.
2.3 Adversarial-Based Active Learning
Adversarial sample-based query strategies have been
used in previous studies. Several researchers (Co-
hen et al., 2021; Thiagarajan et al., 2022; Xia et al.,
2022) use GANs to generate adversarial samples to
facilitate the AL process. However, GAN-based ad-
versarial methods have several limitations. Firstly,
they require additional model training besides the AL
model. Secondly, GANs have a great demand on the
input data to help discriminators learn the distribution
of the data. It conflicts with the primary objective
of AL which aims to complete the training with as
few labels as possible. Thirdly, GANs may generate
images with much noise and sometimes even cannot
be recognized manually (Mayer and Timofte, 2020).
Although several problems above have already been
solved in some cases like MNIST or natural images,
there still are significant challenges in complex med-
ical settings. One would prefer a simple implementa-
tion method that can work with any existing classifier.
DFAL (Ducoffe and Precioso, 2018), as men-
tioned in Section. 1, uses the information provided by
these adversarial examples near the decision bound-
ary on the spatial input distribution to approximate
their distance to the decision boundary, offering a
more efficient way for adversarial-based AL.
3 PROPOSED METHOD
In this section, we start by describing our query strat-
egy for image classification tasks. Then we present
strategies build on this, including two data expansion
methods and adaptive learning, as shown in Fig. 1.
3.1 Query Strategy
For the query strategy, we focus on the samples close
to the decision boundary. The intuition behind it is
straightforward: when a model performs a classifica-
tion task in a high-dimensional space, the instances
close to its decision boundary turn to be highly uncer-
tain. As mentioned in sections 2.2, adversarial attacks
were designed to approximate the slightest perturba-
Adaptive Adversarial Samples Based Active Learning for Medical Image Classification
753
tion to cross decision boundaries, which meets our
needs. As a core part of AL, we adopt the efficient
DeepFool attack, literally computing the smallest per-
turbation for a given image. For a given x ∈ R
m
image
and target label l ∈
{
1...k
}
, the goal is to compute an
additive perturbation ρ ∈ R
m
that would distort the
image very slightly to fool the network:
min||ρ||
2
s.t. h(x +ρ) = l ; x + ρ ∈ [0, 1]
m
(1)
For every unlabeled sample, an overall introduced
perturbation will be recorded and used to sort samples
in the query phase. Then select unlabeled samples
with the smallest adversarial perturbation, i.e., clos-
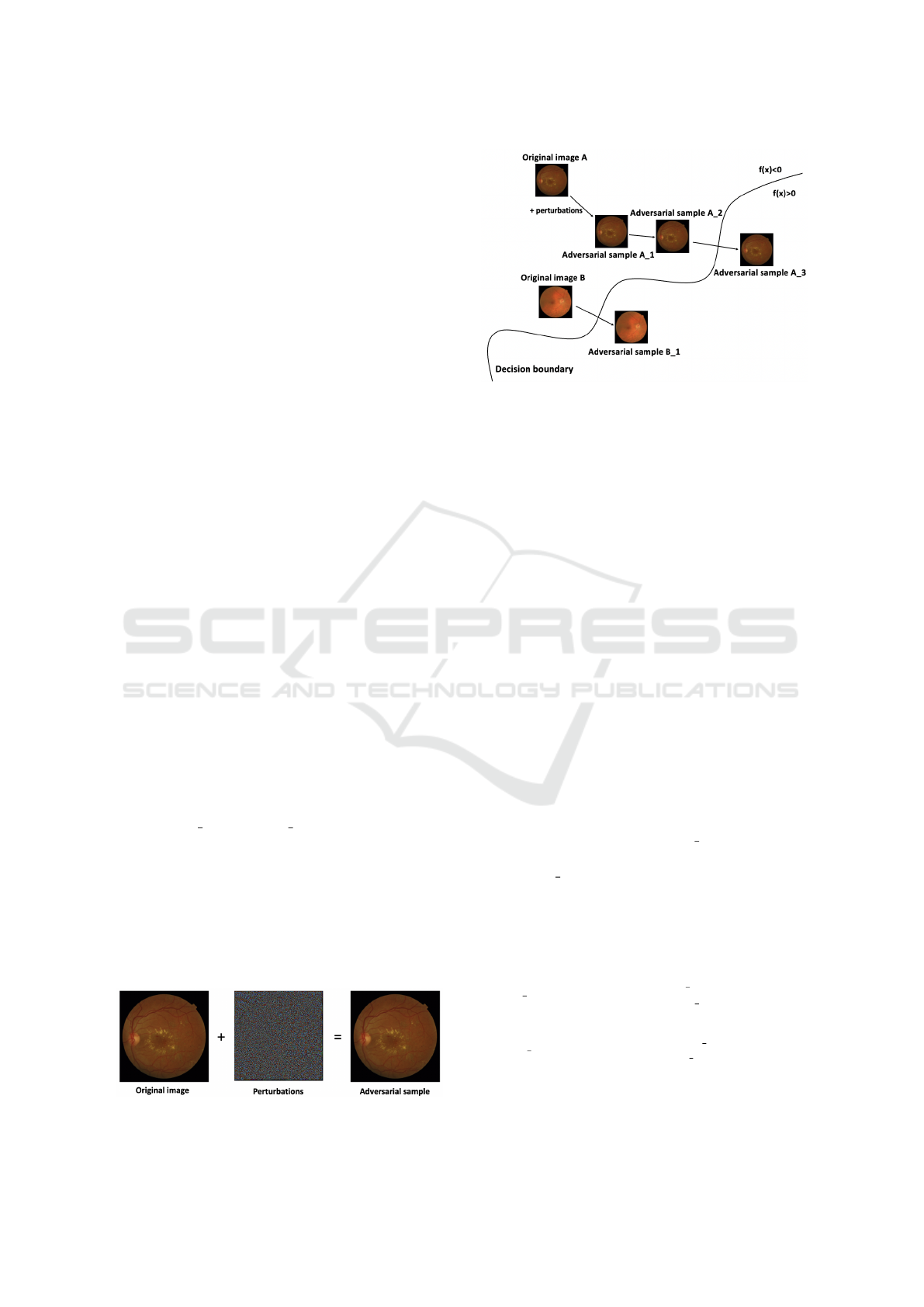
est to the decision boundary to request labels. To il-
lustrate, in Fig. 3, compared to image B, A is more
uncertain, which may need a manual label.
3.2 Adversarial Samples Expansion
During the AL process, adversarial attack gener-
ates counterfactual samples that are indistinguishable
from the original image from a human perspective and
will still belong into the initial category. An example
of a clean image and its corresponding adversarial ex-
ample can be seen in Fig. 2. These generated samples
are even more instructive than the original ones be-
cause they are closer to the current decision boundary
than the original ones but do not require the complex
training process of other networks, such as GANs. As
introduced in section 2.2, medical images are more
vulnerable to adversarial samples. Therefore, as an
extension, we add these generated adversarial sam-
ples (fake images) of uncertain samples to the training
set. These samples may help the model learn essential
features causally related to the pairwise outcomes and
increase model robustness. To clarify, in Fig. 3, after
selecting image A as an uncertain instance, its adver-
sarial sample 2 and sample 3 can be the most valu-
able adversarial sample for augmentation since they
are closest to the decision boundary.
3.3 Pseudo Labeling Expansion
In contrast to data requiring manual labels, some un-
labeled data need larger perturbations to shift the pre-
dictions (image B in Fig. 3), indicating such data have
Figure 2: Adversarial samples of Messidor dataset.
Figure 3: The process of adversarial sampling based on ad-
versarial attack.
high confidence in the prediction. These data are fully
compatible with the most traditional pseudo labeling
method (Dong-Hyun, 2013): unlabeled samples and
their corresponding pseudo labels (model predictions)
are retained for inclusion in the training dataset only
when the classifier has sufficient confidence. There-
fore, we pseudo-annotate them to further reduce the
total amount of manual annotation.
3.4 Adaptive Active Learning
We noticed that the number of samples requested
in each round of the AL is a constant in previous
works. However, from the model’s view, the learn-
ing ability and performance of the models are dif-
ferent in each training round. Therefore, we intro-
duce an adaptive approach to dynamically select sam-
ples at different statuses to utilize data in a more ef-
ficient way. Intuitively, as more training rounds are
completed, the performance of the network on the
training set improves, and it is the same for the test
set if there is no overfitting. In this process, the de-
mand for uncertain samples active number gradually
decreases, while the number of low-uncertainty sam-
ples pseudo number is oppositely increasing. So we
set the number of rounds as a criterion to follow the
exponential function (at an increasing rate) for the re-
quests. Formally speaking, in each iterative training
step, the query number is defined as below:
active number =
max, i f active number < max
a–e
η∗rd
, i f active number ≥ max
(2)
pseudo number =
b + e
η∗rd
, i f pseudo number ≤ min
min, i f pseudo number > min
(3)
where rd stands for training round, a and b are
the start number of labels required and pseudo label,
max and min are the lower bound and upper bound of
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
754

labels required and pseudo label, respectively, and η
represents the changing rate of the whole process.
4 EXPERIMENT
We conducted experiments on two medical datasets
with different features. An introduction of datasets
is given, followed by the description of the imple-
mentation details, including data preprocessing and
augmentation, backbone model, and hyperparame-
ter settings. Then, we show the performance of
our sampling method on two datasets compared to
other benchmarks. Finally, we list a series of abla-
tion studies to demonstrate the usefulness of our pro-
posed components (incorporation of adversarial sam-
ples, pseudo-labeling, and adaptive methods).
4.1 Dataset Description
To verify whether our method works efficiently re-
gardless of tasks, we choose diverse image classifi-
cation tasks. Each of these datasets presents different
learning challenges: Messidor is a binary task, and the
Breast Cancer dataset is a multi-class classification.
Messidor Dataset (Decenci
`
ere et al., 2014): contains
1200 eye fundus images from 654 diabetic and 546
healthy patients. This dataset was labeled for Di-
abetic Retinopathy (DR) grade and risk of macular
edema. In our work, Messidor is used to classify fun-
dus images as healthy (DR grade=0) or diseased (DR
grade>0).
Breast Cancer Diagnosis Dataset (Aresta et al.,
2019): consists of 400 high-resolution histopathology
images of breast tissue cells that are evenly split into
four classes: Normal, Benign, in-situ carcinoma, and
invasive carcinoma (100 images per class) for solving
a multi-classification problem.
4.2 Implementation Details
Data Preprocessing and Augmentation. The im-
age size for all datasets is set to 512 × 512 pixels.
Online data augmentation was used during training
to increase the diversity of data, including 15
◦
ran-
dom rotation, random scaling in the range [0.9, 1],
and random horizontal flips, which were made con-
sistent to paper (Smailagic et al., 2018). Since we
used the model pre-trained on the ImageNet dataset,
we did normalization with ImageNet’s mean and stan-
dard deviation. Table 1 shows the detailed implemen-
tation of datasets. For further implementation, code
(Huang, 2021) is released publicly, including the re-
production of some comparative methods.
Table 1: Implementation details of the data sets, including
the division of each data set, the initialization of the training
set, and the number of images added at each iteration.
Hyper Dataset
Parameters Messidor Breast Cancer
train size 768 320
test size 192 80
Initial training set size 100 30
Images labeled in each cycle 20 10
Backbone Models. We employed Inception V3 to
classify the images in the AL process, with the cross-
entropy loss minimized through supervised label in-
formation. The last layer of the Inception V3 was re-
moved, and the fully-connected layer was added to
achieve the number of output classes we want.
Hyper Parameters. For a fair comparison, we
keep the hyper-parameter settings consistent as pa-
per (Smailagic et al., 2018) for all experiments in this
paper. We used an Adam optimizer with a learning
rate of 0.0002 and weight decay of 0.01. We set the
batch size to 8 and the maximum epoch to 300. At
each AL iteration, the model is trained until obtaining
100% accuracy on the training set. The model’s pa-
rameters were reset to the pre-trained weights from
ImageNet after each iteration, while the new fully-
connected layer was initialized with random weights
using the glorot method (Glorot and Bengio, 2010).
Evaluation Matrix. We use accuracy as the perfor-
mance evaluation matrix for both datasets. To eval-
uate the AL process, we look at two aspects: the
minimum number of manually labeled data used to
achieve the same performance as supervised learning
and the highest performance achieved in the process.
4.3 Experimental Results
As shown in Table 2, our method without adaption
(combining only adversarial samples expansion with
pseudo labels) and our method with adaption (with
adaptive learning module), achieved consistently su-
perior performance compared to the seven baselines
on the test set of the Messidor dataset. Specifically,
our method uses only 260 (33.9%) and 295 (38.4%)
images to reach the supervised baseline, respectively,
compared to 580 (75.5%) images of the DeepFool
method. In particular, the proposed adaptive method
exceeded the supervised baseline in testing accuracy
(0.8667) and peaked at 0.9.
For the Breast Cancer Diagnosis dataset, our
methods used less labeled data (no more than 50%
data) to reach the supervised baseline compared to
other methods. The method’s accuracy with adaption
is 0.925, which is 7% higher than supervised learning
performance. Table 3 shows the result.
Adaptive Adversarial Samples Based Active Learning for Medical Image Classification
755

Table 2: Performance comparison on the Messidor dataset.
Query Number of labeled data Number of labeled data to Highest
Methods 200 300 400 500 600 700 All achieve supervised baseline accuracy
Random selection 0.6917 0.7583 0.6958 0.7750 0.8042 0.8708 0.8833 700 0.8833
Entropy 0.7125 0.7292 0.8334 0.8792 0.8458 0.8917 0.8667 500 0.8917
MC Dropout 0.7333 0.775 0.8125 0.8125 0.8458 0.8500 0.8292 620 0.8708
BALD 0.7208 0.7708 0.8000 0.8417 0.8417 0.8375 0.8417 620 0.8917
DeepFool 0.7542 0.7667 0.8125 0.8125 0.8334 0.8667 0.8458 580 0.8792
MedAL 0.8042 0.8417 0.8217 0.7042 0.8375 0.7333 0.8333 / 0.8542
OMedAL 0.8208 0.8500 0.8542 0.8917 0.8417 0.8792 0.8417 420 0.8917
Our method without adaption 0.7667 0.8417 0.8667 / / / / 260 0.8750
Our method with adaption 0.8 (184) 0.875 (295) 0.8708 (402) 0.8968 (504) / / / 295 0.8958
Table 3: Performance comparison on the Breast Cancer diagnosis dataset.
Query Number of labeled data Number of labeled data to Highest
Methods 50 100 150 200 250 300 All achieve supervised baseline accuracy
Random selection 0.7250 0.7000 0.7625 0.8125 0.8500 0.8500 0.8750 310 0.8750
Entropy 0.6375 0.7250 0.8000 0.8625 0.8875 0.8750 0.8375 190 0.8875
MC Dropout 0.6750 0.7125 0.8500 0.8500 0.8500 0.8875 0.8625 220 0.9000
BALD 0.7000 0.7250 0.8000 0.8250 0.8750 0.8500 0.8250 230 0.8875
DeepFool 0.7000 0.7875 0.8125 0.8375 0.9000 0.8250 0.8625 190 0.9000
MedAL 0.5875 0.5875 0.6250 0.8000 0.8875 0.9250 0.9500 230 0.9500
OMedAL 0.8250 0.8000 0.8250 0.8125 0.7875 0.8375 0.8875 160 0.9000
Our method without adaption 0.775 0.825 0.8375 / / / / 160 0.8750
Our method with adaption 0.675 (48) 0.825 (102) 0.8625 (155) 0.875 (206) / / / 155 0.9250
Overall, while the performance between Deep-
Fool and other baselines is similar, our method shows
consistently superior performance in different scenar-
ios. It is worth noting that in this process, we use
fewer rounds to achieve better results than our main
comparator (the DeepFool method), proving the ef-
fectiveness of our method.
4.4 Ablation Studies
To verify the effectiveness of our method, we take
DeepFool-based AL (Ducoffe and Precioso, 2018) as
the main comparison for the subsequent sections. We
evaluate the method by monitoring the test accuracy
after each AL iteration.
4.4.1 Adversarial Samples Expansion
We experimented with the counterfactual images nat-
urally generated by adversarial attack. We com-
pare performance with or without (DeepFool) adding
adversarial samples, and the numbers of adversar-
ial samples: with one (DeepFool add1: the one
that ended up being wrongly classified), two (Deep-
Fool add2: about to be wrongly scored the one al-
ready been wrongly scored, e.g., the adversarial sam-
ple 2 and sample 3 in Fig. 3) for data with high uncer-
tainty or all the adversarial samples generated during
the process (DeepFool addall), to analyze the effect
of adding adversarial samples and the total number in
this AL process.
As illustrated in Fig. 4, the result shows the advan-
tage of adversarial sample expansion, which improves
the final model effect. For DeepFool add1, perfor-
mance on the test set is on average 5% higher per
Figure 4: Performance of counterfactual expansion on AL.
round when adding one adversarial sample with every
uncertain data, which is more pronounced in the mid-
dle process. Finally, only 360 samples are needed to
achieve comparable performance. However, adding
two or more samples exacerbated the instability of the
results, although DeepFool add2 achieved the base-
line score with only 480 samples and finally reached
0.9125, which is the best result of all these experi-
ments in this respect. Therefore, we believe that ad-
versarial samples increase the model’s generalizabil-
ity, but the number of adversarial samples added in
each round should be chosen carefully: as the number
of adversarial samples increases, the performance be-
comes progressively more unstable and worse, which
we speculate is due to the effect of the noise intro-
duced by the adversarial samples.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
756
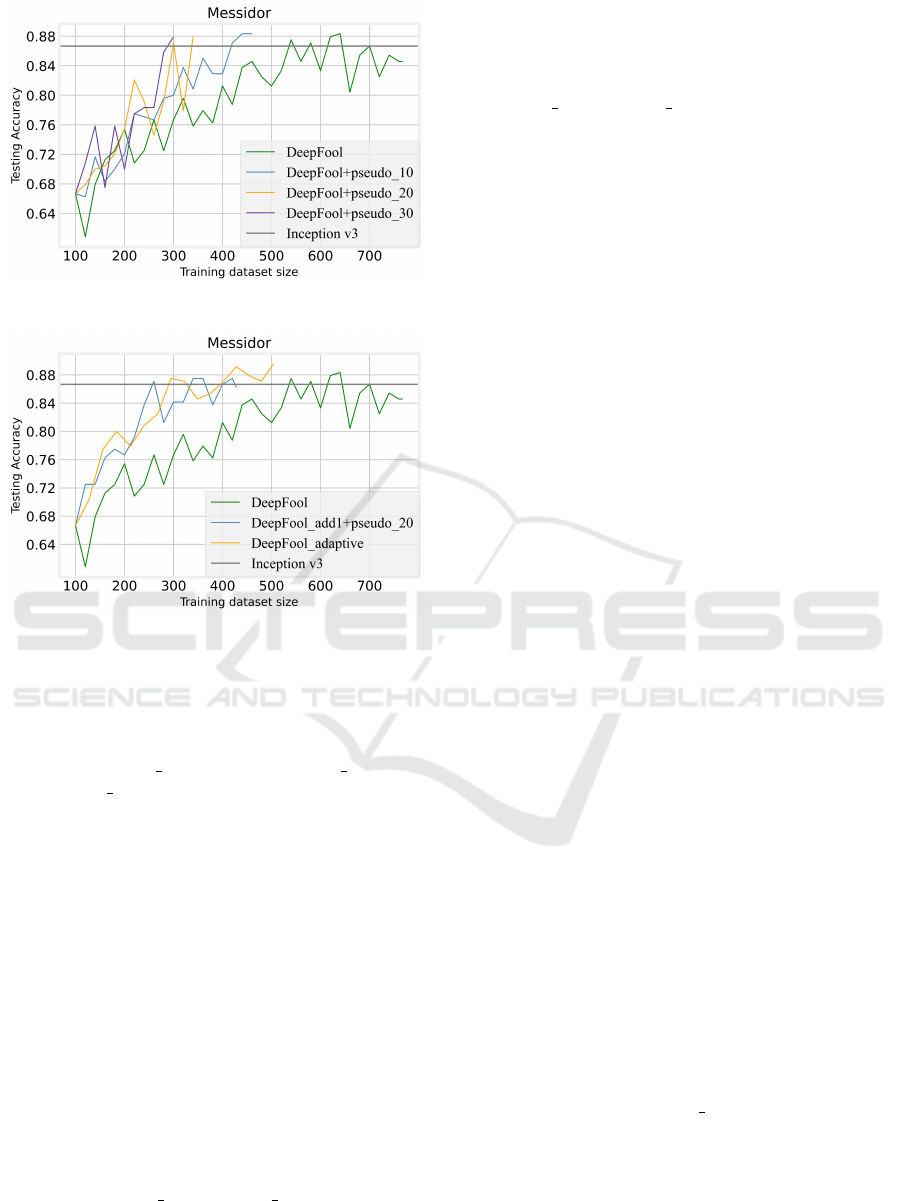
Figure 5: Experiment results of pseudo labels on AL.
Figure 6: The experiment result of adaptive AL.
4.4.2 Pseudo Label Expansion
In this part, we conducted experiments on the
pseudo labeling, adding 10, 20, and 30 samples
(DeepFool+pseudo 10, DeepFool+pseudo 20, Deep-
Fool+pseudo 30) furthest from the decision boundary
in each round, respectively, to analyze the effect of
pseudo labels on the AL process.
From Fig. 5, when 10, 20, and 30 pseudo labels
are added in each round, it reached the supervised
learning baseline with only 420, 300, and 280 labels,
respectively, compared with 540 labels in DeepFool.
Therefore, we conclude that involving pseudo labels
can effectively reduce the burden of manual labeling
and save AL time by reducing the number of selec-
tion rounds, even if some fluctuations are brought as
the number of pseudo labeling increases.
4.4.3 Adaptive Active Learning
In this experiment, we first synthesized the combina-
tion of the two expansion methods and then experi-
mented with adaptive AL based on this.
Line DeepFool
add1+pseudo 20 illustrates that
when we combine the two previous data expansion
skills, less than 260 images are needed to achieve the
fully supervised performance, compared to 530 im-
ages for the pure DeepFool method. Furthermore,
the proposed adaptive method also shows its superi-
ority: while reaching the baseline similarly to method
DeepFool add1+pseudo 20, it achieves an higher ac-
curacy of nearly 0.9, compared to 0.8667 of the super-
vised baseline. Correspond to the mathematical ex-
pressions in 3.4, the experiment hyperparameter set-
ting for adaptive learning are: η, max, min are set to
0.1, 20 and 5 for both datasets, while a is 30 and 20, b
is 15 and 5 respectively for Messidor and Breast Can-
cer datasets.
5 DISCUSSION
One observable phenomenon is that the test accuracy
sometimes has jitters in each round, which we con-
jecture is partly due to the selection and fitting of
the model itself. However, for the large fluctuations
(whether drops or rises), it is worth exploring the rea-
sons in subsequent work to discover what features
motivate the plunges from the data and model level.
Although our method has already improved the ef-
fectiveness of data utilization skills, efficiency should
be improved, especially for real-world implementa-
tion. Therefore, we plan to propose some computa-
tional efficiency methods for each round by retraining
and stopping strategies.
6 CONCLUSIONS
In this work, we proposed a new adversarial AL im-
plementation for medical image classification tasks
using only a few annotations. It demonstrates the
validity of data utilization skills and adaptive selec-
tion in AL, which outperforms multiple state-of-the-
art and exceeds the supervised baseline in terms of
final results. We also demonstrate the robustness of
our approach by conducting experiments on medical
datasets with different features.
ACKNOWLEDGEMENTS
This work was supported by Science Foundation Ire-
land (SFI) [SFI/12/RC/2289 P2].
REFERENCES
Aresta, G., Ara
´
ujo, T., Kwok, S., Chennamsetty, S. S.,
Safwan, M., Alex, V., Marami, B., Prastawa, M.,
Adaptive Adversarial Samples Based Active Learning for Medical Image Classification
757

Chan, M., Donovan, M., et al. (2019). Bach: Grand
challenge on breast cancer histology images. Medical
image analysis, 56:122–139.
Ash, J. T., Zhang, C., Krishnamurthy, A., Langford, J., and
Agarwal, A. (2019). Deep batch active learning by di-
verse, uncertain gradient lower bounds. arXiv preprint
arXiv:1906.03671.
Cohen, J. P., Brooks, R., En, S., Zucker, E., Pareek, A.,
Lungren, M. P., and Chaudhari, A. (2021). Gifsplana-
tion via latent shift: a simple autoencoder approach to
counterfactual generation for chest x-rays. In Medical
Imaging with Deep Learning, pages 74–104. PMLR.
Decenci
`
ere, E., Zhang, X., Cazuguel, G., Lay, B., Coch-
ener, B., Trone, C., Gain, P., Ordonez, R., Massin, P.,
Erginay, A., et al. (2014). Feedback on a publicly dis-
tributed image database: the messidor database. Im-
age Analysis & Stereology, 33(3):231–234.
Dong-Hyun, L. (2013). Pseudo-label: The simple and effi-
cient semi-supervised learning method for deep neural
networks. In Workshop on challenges in representa-
tion learning, ICML, volume 3, page 896.
Ducoffe, M. and Precioso, F. (2018). Adversarial active
learning for deep networks: a margin based approach.
arXiv preprint arXiv:1802.09841.
Finlayson, S. G., Bowers, J. D., Ito, J., Zittrain, J. L., Beam,
A. L., and Kohane, I. S. (2019). Adversarial attacks on
medical machine learning. Science, 363(6433):1287–
1289.
Gal, Y., Islam, R., and Ghahramani, Z. (2017). Deep
bayesian active learning with image data. In Interna-
tional Conference on Machine Learning, pages 1183–
1192. PMLR.
Glorot, X. and Bengio, Y. (2010). Understanding the diffi-
culty of training deep feedforward neural networks. In
Proceedings of the thirteenth international conference
on artificial intelligence and statistics, pages 249–
256. JMLR Workshop and Conference Proceedings.
Houlsby, N., Husz
´
ar, F., Ghahramani, Z., and Lengyel, M.
(2011). Bayesian active learning for classification and
preference learning. arXiv preprint arXiv:1112.5745.
Huang, K.-H. (2021). Deepal: Deep active learning in
python. arXiv preprint arXiv:2111.15258.
Joshi, A. J., Porikli, F., and Papanikolopoulos, N. (2009).
Multi-class active learning for image classification. In
2009 ieee conference on computer vision and pattern
recognition, pages 2372–2379. IEEE.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., Van Der Laak, J. A.,
Van Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
image analysis, 42:60–88.
Ma, X., Niu, Y., Gu, L., Wang, Y., Zhao, Y., Bailey, J., and
Lu, F. (2021). Understanding adversarial attacks on
deep learning based medical image analysis systems.
Pattern Recognition, 110:107332.
Mayer, C. and Timofte, R. (2020). Adversarial sampling
for active learning. In Proceedings of the IEEE/CVF
Winter Conference on Applications of Computer Vi-
sion, pages 3071–3079.
Paschali, M., Conjeti, S., Navarro, F., and Navab, N. (2018).
Generalizability vs. robustness: investigating medical
imaging networks using adversarial examples. In In-
ternational Conference on Medical Image Computing
and Computer-Assisted Intervention, pages 493–501.
Springer.
Pervin, M., Tao, L., Huq, A., He, Z., Huo, L., et al. (2021).
Adversarial attack driven data augmentation for accu-
rate and robust medical image segmentation. arXiv
preprint arXiv:2105.12106.
Ren, X., Zhang, L., Wei, D., Shen, D., and Wang, Q. (2019).
Brain mr image segmentation in small dataset with ad-
versarial defense and task reorganization. In Inter-
national Workshop on Machine Learning in Medical
Imaging, pages 1–8. Springer.
Sener, O. and Savarese, S. (2017). Active learning for
convolutional neural networks: A core-set approach.
arXiv preprint arXiv:1708.00489.
Settles, B. (2009). Active learning literature survey.
Sinha, S., Ebrahimi, S., and Darrell, T. (2019). Varia-
tional adversarial active learning. In Proceedings of
the IEEE/CVF International Conference on Computer
Vision, pages 5972–5981.
Smailagic, A., Costa, P., Gaudio, A., Khandelwal, K., Mir-
shekari, M., Fagert, J., Walawalkar, D., Xu, S., Gal-
dran, A., Zhang, P., et al. (2020). O-medal: Online ac-
tive deep learning for medical image analysis. Wiley
Interdisciplinary Reviews: Data Mining and Knowl-
edge Discovery, 10(4):e1353.
Smailagic, A., Costa, P., Noh, H. Y., Walawalkar, D., Khan-
delwal, K., Galdran, A., Mirshekari, M., Fagert, J.,
Xu, S., Zhang, P., et al. (2018). Medal: Accurate and
robust deep active learning for medical image analy-
sis. In 2018 17th IEEE international conference on
machine learning and applications (ICMLA), pages
481–488. IEEE.
Szegedy, C., Zaremba, W., Sutskever, I., Bruna, J., Er-
han, D., Goodfellow, I., and Fergus, R. (2013). In-
triguing properties of neural networks. arXiv preprint
arXiv:1312.6199.
Thiagarajan, J. J., Thopalli, K., Rajan, D., and Turaga, P.
(2022). Training calibration-based counterfactual ex-
plainers for deep learning models in medical image
analysis. Scientific Reports, 12(1):1–15.
Tran, T., Do, T.-T., Reid, I., and Carneiro, G. (2019).
Bayesian generative active deep learning. In Interna-
tional Conference on Machine Learning, pages 6295–
6304. PMLR.
Xia, T., Sanchez, P., Qin, C., and Tsaftaris, S. A. (2022).
Adversarial counterfactual augmentation: Application
in alzheimer’s disease classification. arXiv preprint
arXiv:2203.07815.
Zhdanov, F. (2019). Diverse mini-batch active learning.
arXiv preprint arXiv:1901.05954.
Zhu, J.-J. and Bento, J. (2017). Generative adversarial ac-
tive learning. arXiv preprint arXiv:1702.07956.
ICPRAM 2023 - 12th International Conference on Pattern Recognition Applications and Methods
758
