
Deep Hybrid Bagging Ensembles for Classifying Histopathological
Breast Cancer Images
Fatima-Zahrae Nakach
1
, Ali Idri
1,2
and Hasnae Zerouaoui
1
1
Modeling, Simulation and Data Analysis, Mohammed VI Polytechnic University, Marrakech-Rhamna, Benguerir, Morocco
2
Software Project Management Research Team, ENSIAS, Mohammed V University, Rabat-Salé-Kénitra, Rabat, Morocco
Keywords: Ensemble Learning, Bagging, Transfer Learning, Breast Cancer.
Abstract: This paper proposes the use of transfer learning and ensemble learning for binary classification of breast
cancer histological images over the four magnification factors of the BreakHis dataset: 40×, 100×, 200× and
400×. The proposed bagging ensembles are implemented using a set of hybrid architectures that combine pre-
trained deep learning techniques for feature extraction with machine learning classifiers as base learners (MLP,
SVM and KNN). The study evaluated and compared: (1) bagging ensembles with their base learners, (2)
bagging ensembles with a different number of base learners (3, 5, 7 and 9), (3) single classifiers with the best
bagging ensembles, and (4) best bagging ensembles of each feature extractor and magnification factor. The
best cluster of the outperforming models was chosen using the Scott Knott (SK) statistical test, and the top
models were ranked using the Borda Count voting system. The best bagging ensemble achieved a mean
accuracy value of 93.98%, and was constructed using 3 base learners, 200× as a magnification factor, MLP
as a classifier, and DenseNet201 as a feature extractor. The results demonstrated that bagging hybrid deep
learning is an effective and a promising approach for the automatic classification of histopathological breast
cancer images.
1 INTRODUCTION
Breast cancer (BC) became the most commonly
diagnosed cancer type in the world in 2021, impacting
more than 2.1 million women and causing more than
one million deaths per year (Sung et al. 2021). The
early detecting and accurate diagnosis of BC can
improve the survival rate of BC patients (Clegg et al.
2009). Histopathology biopsy imaging is currently
the gold standard for identifying BC in clinical
practice since it provides a comprehensive view of
how the malignancy affects the tissues (Kumar et al.
2017). However, it is difficult to categorize some
intricate visual patterns as benign or malignant in the
histopathology slides (Gupta and Bhavsar 2017).
Machine learning (ML) models can assist
pathologists for automatic cancer disease detection by
improving the accuracy and speeding up the diagnosis
process (din et al. 2022). The identification of the
tumor type is a necessary step before a patient can be
given a better treatment plan that increases their
chance of survival, that is why ML models have to
initially distinguish benign from malignant tumors
(Kumar et al. 2017). Several ML classifiers have been
applied in cancer research for the development of
predictive models (Saxena and Gyanchandani 2020),
but none of them has been proved to always
outperform the others (Zerouaoui and Idri 2021;
Nemade et al. 2022).
To avoid making a poor predictive decision based
on a single selected model (Shen et al. 2020), ensemble
learning aims to improve the performance of one
algorithm by using an intelligent combination of
several individual models (also called base learners)
and reducing both variance and bias (Tuv 2006). In
fact, previous research has shown that an ensemble is
often more accurate than any of the base learners
(Hastie et al. 2009). However, it must be considered
that in some cases the best individual classification
model within the ensemble might beat the performance
of its ensemble (Polikar 2012). Bagging is an ensemble
learning method based on the aggregation of the
decision of different predictors that were parallelly
trained on different random subsets of the dataset using
the same ML algorithm (Breiman 1996).
To address the BC binary classification of
histopathological images, researchers have used
different ensemble techniques that outperform single
Nakach, F., Idri, A. and Zerouaoui, H.
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images.
DOI: 10.5220/0011704200003393
In Proceedings of the 15th International Conference on Agents and Artificial Intelligence (ICAART 2023) - Volume 2, pages 289-300
ISBN: 978-989-758-623-1; ISSN: 2184-433X
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
289

classifiers (Abdar and Makarenkov 2019; Abdar et al.
2020; Hameed et al. 2020; Nakach et al. 2022a;
Alaoui et al. 2022; Abbasniya et al. 2022). For
bagging ensembles, the majority of papers are
classifying the tumors using tabular data:
(Nascimento et al. 2011; Wang et al. 2018; Naveen et
al. 2019; Sharma and Deshpande 2021), BC
mammography images (Ponnaganti and Anitha 2022)
(Lbachir et al. 2021) and gene expression (Otoom et
al. 2015; Wu and Hicks 2021) or they are using the
bagging ensemble to compare it with the performance
of another proposed method (Yadavendra and Chand
2020). For histopathological images, this paper
(Zhang et al. 2013) used traditional ML classifiers
with bagging as Support Vector Machine (SVM),
Multilayer Perceptron (MLP), and Decision Trees
(DT) without CNNs and without varying the number
of base learners. On the other hand, the papers that
used a hybrid architecture only used DT as a base
learner (Guo et al. 2018) (Nakach et al. 2022b).
In order to confirm or refute the usefulness of
bagging ensembles in diagnosing BC in comparison
with single ML classifiers, the present study develops
and evaluates a set of bagging hybrid architecture
ensembles for binary classification of BC
histopathological images over the BreakHis dataset at
different magnification factors (MFs): 40×, 100×,
200×, and 400×. The forty-eight bagging ensembles
are designed using: (1) three of the most recent DL
techniques for feature extraction using transfer
learning: DenseNet201, MobileNetV2, and
InceptionV3, (2) three of the most popular ML
classifiers as base learners: MLP, K-Nearest
Neighbours (KNN), and SVM, and (3) different
number of base learners (3, 5, 7 and 9 estimators).
The main objective is to look into the effect of the
type and number of base learners on the predictive
capability of the bagging ensembles, and to examine
the impact of various feature extraction DL models
on the performance of the bagging ensembles. For
that purpose, four research questions (RQs) are
explored in this study:
• (RQ1). Do Bagging Ensembles Outperform their
base Learners?
• (RQ2). How the Number of base Learners
Impacts the Performance of Bagging Ensembles?
• (RQ3). Do Bagging Ensembles Outperform
Their Single Classifiers?
• (RQ4). Is There any Bagging Ensemble that
Outperforms the Others for Each MF and Over all
the MFs?
The rest of the paper is as follows. A summary of
related work is given in Section 2. Section 3 presents
data preparation and the empirical methodology
followed throughout the research. Section 4 reports
the empirical findings and discussions. Section 5
highlights the conclusion and future direction of this
study.
2 RELATED WORK
This section presents an overview of primary studies
investigating ensemble learning for BC
histopathological image classification. Kassani et al.
(Kassani et al. 2019) created a bagging ensemble with
pre-trained CNNs: MobileNetV2, VGG19 and
DenseNet201 for automatic binary classification of
breast histopathological images, the results proved
that the best single classifier is DenseNet201 and that
the proposed multi-model ensemble method obtains
better predictions than single classifiers of VGG19,
MobileNetV2, and DenseNet201. Additionally, the
proposed ensemble outperforms ML algorithms (DT,
Random Forest, XGBoost, AdaBoost and Bagging
Classifier) with an accuracy of 98.13%, for BreakHis
dataset (MF independent). For ML algorithms and the
BreakHis dataset, the topmost result was obtained by:
the bagging ensemble with 94.97% accuracy,
XGBoost with an accuracy of 94.11%, followed by
Random Forest with an accuracy of 92.10%, and
AdaBoost with an accuracy of 91.82%.
In another study (Zhu et al. 2019), the bagging
ensembles were constructed using hybrid CNN
models to classify the BreakHis dataset with its 4 MFs
and the BACH dataset. The results showed that
combining multiple compact CNNs led to an
improvement in classification performance, with an
accuracy of 85.7%, 84.2%, 84.9% and 80.1% for MF
40×, 100×, 200× and 400× respectively.
Additionally, Wang et al. (Wang et al. 2020)
designed a bagging DT ensemble and a subspace
discriminant classifiers ensemble to classify the
images of the BreakHis dataset, the DT bagging
ensemble achieved the highest accuracy rate of 89.7%
compared to the single classifiers. For multi-
classification, the subspace discriminant classifier
ensemble gave the accuracy of 88.1%. Moreover, in
(Alaoui et al. 2022), deep stacked ensembles were
developed using seven pre-trained deep learning (DL)
models: VGG16, VGG19, ResNet 50, InceptionV3,
Inception ResNet V2, Xception, and MobileNet V2,
then a logistic regression was used as a meta-learner
that learns how to best combine the predictions of the
DL models. The results showed that the proposed
deep stacking ensemble reports an overall accuracy of
93.8%, 93.0%, 93.3%, and 91.8% over the four MF
values of the BreakHis dataset: 40×, 100×, 200× and
400×, respectively.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
290

3 MATERIALS AND METHODS
This section presents: the publicly available dataset
used in the empirical evaluations carried out in this
study, the different performance criteria used to
evaluate the models, the experimental process and the
abbreviations.
3.1 Data Preparation
In this paper the Breast Cancer Histopathological
Image Classification (BreakHis) dataset (Spanhol et
al. 2016) is used in order to examine the viability of
the hybrid bagging ensembles. Before feeding the
input images into the proposed models, the pre-
processing is a crucial step which aims to improve the
quality of their visual information. In this study, the
preprocessing step is similar to the process that has
been followed in (Zerouaoui et al. 2021) and
(Zerouaoui and Idri 2022) where they used intensity
normalization (B 2019) and Contrast Limited
Adaptive Histogram Equalization (CLAHE) (Yussof
2013). After the pre-processing step, data
augmentation (Shorten and Khoshgoftaar 2019) was
used to avoid overfitting and balance the data by
increasing the number of benign images.
3.2 Performance Measures
The performance criteria used to evaluate bagging
ensembles as well as their base learners and single
models are: accuracy, precision, sensitivity and F1-
score. These metrics have been the most frequently
used in classification (Zerouaoui and Idri 2021) and
they are mathematically expressed by the equations
1-4 respectively:
Accuracy=(TP+TN)/(TN+TP+FP+FN)
(1)
Precision=TP/(TP+FP)
(2)
Sensitivity =TP/(TP+FN)
(3)
F1-score=2×(Recall×precision)/(Recall+precision)
(4)
where: TP is a malignant case that is identified as
malignant, FP is a benign case that is identified as
malignant, TN is a benign case that is identified as
benign, and FN is a malignant case that is identified
as benign.
3.3 Experimental Process
The empirical evaluations of the hybrid bagging
ensembles use three classifiers: MLP, SVM and
KNN, and three DL feature extractors (FE):
InceptionV3, DenseNet201, and MobileNetV2, over
the four MFs of the BreakHis dataset: 40×, 100×,
200×, and 400×. To compare the models, this study
used the SK statistical test (Jelihovschi et al. 2014)
based on accuracy to cluster the models and identify
the best SK cluster. The best SK cluster contains one
or many models that have the best accuracy and that
are statistically indifferent. If many models are found,
they will be ranked using the Borda Count voting
system (Emerson 2013) based on the four
performance measures: accuracy, sensitivity,
precision and F1-score. The 5-fold cross validation
was employed to guarantee that every observation
from the original dataset has a chance of appearing in
both the training and test set and that the images
selected for testing were not used during training in
order to successfully complete the binary
classification task. The empirical evaluations involve
eight steps as shown in Figure 1. The present study
has the same potential threats to validity of the
approach in this study (Nakach et al. 2022b).
3.4 Abbreviation
The abbreviation of bagging ensembles is EBG, plus
the first letter of the classifier used (K for KNN, M
for MLP and S for SVM) and the first letter of the FE
technique used (D for DenseNet201, I for
InceptionV3 and M for MobileNetV2) followed by
the number of base learners.
In tables and figures, the DL techniques were
abbreviated using: DENSE for DenseNet_201, MOB
for MobileNet_V2, and INC for Inception_V3.
4 RESULTS AND DISCUSSIONS
This section presents and discusses the results of the
empirical evaluations, where each is dedicated for
one RQ.
4.1 Do Bagging Ensembles Perform
Better than Their Base Learners?
This section evaluates and compares the bagging
ensembles with their base learners. For each MF, FE,
and classifier, the bagging ensembles were first
compared in terms of accuracy with their base
learners. For each MF and FE, it was observed that:
-For KNN and MLP, the accuracy of the bagging
ensembles is always better than the mean accuracy of
their base learners.
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images
291

Figure 1: Empirical Design.
- For SVM, the mean accuracy of the base learners
outperformed the accuracy of bagging ensembles
constructed using: 9 base learners with MobileNetV2
as FE and MF 100× and 400×, 3 and 5 base learners
with InceptionV3 as FE and MF 400×. All the other
SVM bagging ensembles have a higher accuracy than
the mean accuracy of their base learners.
-The difference of accuracies varies between:
13.04% and 4.58% for MLP, 5.85% and 0.56% for
KNN, and 1.48% and 0.02% for SVM.
The SK test based on accuracy was carried out
over each MF and FE and for each classifier to check
whether there was a notable difference between the
performance of the bagging ensembles and their base
learners. The results showed that:
• For MLP, only one cluster was obtained over
each MF and FE except for MF 400× with
DenseNet201 and InceptionV3, where the bagging
ensembles significantly outperformed one or two of
their base learners (Figure 2).
• For SVM, only one cluster was identified over
each FE and for MF 100×, 200× and 400×. For MF
40×, always one cluster was obtained except for
InceptionV3 as FE, where the bagging ensembles
with 7 and 9 base learners outperformed one of their
base learners.
• For KNN, only one cluster was obtained over
each MF and FE except for: (1) MobileNetV2 as FE
and MF 200× where the best cluster contains the
bagging ensemble with 3 base learners, and (2)
InceptionV3 and MF 400× where the best cluster
contains the bagging ensemble with 9 base learners,
the second cluster contains all the base learners.
Figure 2: SK test Results of the MLP bagging ensemble
using MF 400×, 9 base learners and DenseNet201 as FE.
The models that belong to the same best cluster
were ranked using the Borda Count voting system:
• For MLP, the bagging ensemble is generally
ranked first whatever the MF and FE are. The bagging
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
292

ensemble was ranked second or third for
DenseNet201 over MF 40× regardless the number of
base learners, and it was ranked second for
InceptionV3 over MF 200× with 7 and 9 base learners
and with MobileNetV2 over MF 40× for 5 base
learners.
• For SVM, the bagging ensembles had the worst
ranks (4th, 5th and sometimes 6th) and only 20
ensembles out of 48 were ranked first.
• For KNN, the bagging ensembles are always
ranked first except for the bagging ensemble
constructed using 7 base learners with KNN and
DenseNet201 over the MF 200×.
To conclude, the bagging ensembles outperform
their base learners for MLP, KNN, but for SVM, the
base learners often perform better than the bagging
ensembles. Besides, the differences of accuracies
between the MLP bagging ensembles and their base
learners were relatively important compared to the
differences of accuracies between the SVM and KNN
bagging ensembles and their base learners, which was
expected since bagging ensembles perform better
with base learners that have a high variance such as
MLP and DT (Tuv 2006)(Opitz and Maclin 1999).
This paper (Nakach et al. 2022b) also found that the
bagging ensembles using DT outperform their base
learners for each MF).
4.2 How the Number of Base Learners
Impacts the Performances of
Bagging Ensembles?
Figures 3–5 show the comparison of the accuracy
values of the different bagging ensembles over each
classifier using the four number of base learners, three
DL techniques as FEs (DenseNet201, InceptionV3
and MobileNetV2) and four MF values: 40×, 100×,
200× and 400×.
For the MLP Classifier:
• The highest accuracy was obtained when using the
DenseNet201 as FE regardless the MF values: 40×,
100×, 200× and 400×, and the lowest accuracy was
obtained using MobileNetV2 for FE.
For the SVM Classifier:
• The highest accuracy was obtained when using the
InceptionV3 as FE with the MF value 40× and 400×,
and with DenseNet201 with the MF value 100× and
200×. The lowest accuracy was obtained using
MobileNetV2 as FE with all MF values.
For the KNN Classifier:
• The highest accuracy was obtained when using the
InceptionV3 as FE for the MF 40×, and with
DenseNet201 for the MF 100× and 200×, and with
MobileNetV2 for MF 400×. The lowest accuracy was
obtained when using InceptionV3 regardless the MF
values.
Thereafter, the SK statistical test was used to
cluster the bagging ensembles with different number
of base learners for each classifier, FE and MF. For
MLP and SVM, the SK test of the bagging ensembles
contains one cluster over each FE and MF, which
implies that regardless the number of base learners
used the accuracy of the ensembles is statistically
similar. For KNN, the SK test identified one cluster
with all the bagging ensembles for all the MFs and
FEs except for InceptionV3 over the MF 100×, where
two clusters were identified and the worst one
contains the ensemble of 9 base learners. Table 1
displays the ranking provided by the Borda Count
voting system for the bagging ensembles over each
classifier, FE and MF: the first number represents the
number of base learners associated to the best
bagging ensemble.
It was found that regardless the hybrid
architecture used, the ensemble of 9 base learners
represents the best ensemble for the majority of
bagging ensembles (31 from 48) and it was ranked
last only 3 times (SVM with MF 40× and InceptionV3
and with MF 100× and MobileNetV2, and for KNN
with MF 40× and MobileNetV2). Contrarily, the
ensemble of 3 base learners was only ranked twice as
the best ensemble and 33 times as the worst one.
The bagging ensemble of 5 and 7 base learners
were ranked first 8 and 7 times out of 48 respectively.
Table 1: Borda Count ranking of the bagging ensembles of
each classifier.
MF FE MLP SVM KNN
40×
DENSE
3-9-5-7
9-5-7-3 9-7-5-3
MOB
9-7-5-3
7-9-5-3 5-7-3-9
INC
9-7-5-3
7-5-3-9 9-7-5-3
100×
DENSE
9-5-7-3
9-7-3-5 9-5-7-3
MOB
9-3-7-5
5-7-3-9 9-7-5-3
INC
7-9-5-3
7-9-5-3 9-7-5-3
200×
DENSE
9-7-3-5
5-3-9-7 3-5-9-7
MOB
9-5-7-3
9-7-5-3 5-9-3-7
INC
5-7-9-3
5-9-3-7 9-7-5-3
400×
DENSE
9-7-3-5
9-3-7-5 7-5-9-3
MOB
9-7-3-5
5-7-9-3 5-9-7-3
INC
9-7-3-5
9-7-5-3 9-7-5-3
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images
293
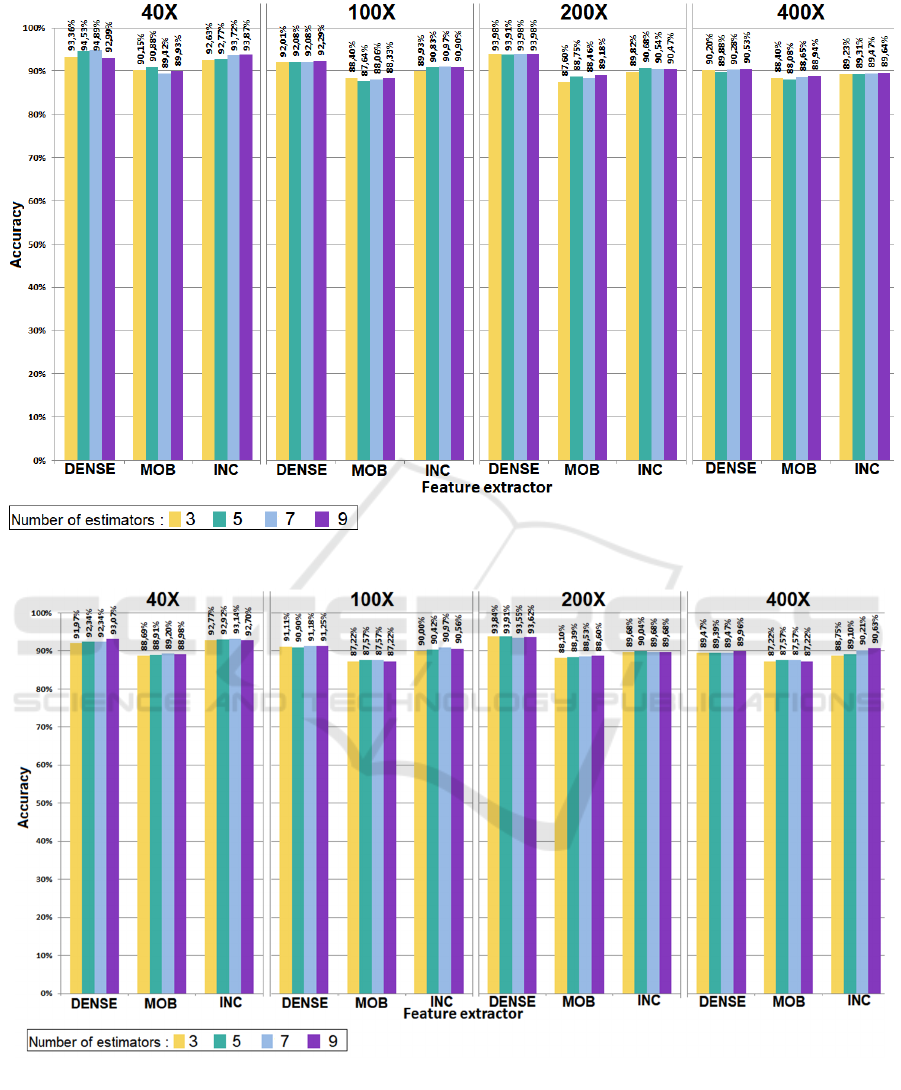
Figure 3: Accuracy values of the bagging ensembles using MLP Classifier over each MF and FE.
Figure 4: Accuracy values of the bagging ensembles using SVM Classifier over each MF and FE.
It can be concluded that, for the majority of MFs
and FEs, the bagging ensembles designed with KNN
and MLP and 9 base learners were the best ensemble
while those designed with 3 base learners represents
the worst ones. For SVM, the bagging ensembles did
not seem to respect a specific order.
4.3 Do Bagging Ensembles Perform
Better than Single Classifiers?
This section compares the single model and the best
bagging ensemble of each classifier. Tables 2-4
summarize the testing accuracy values of the single
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
294

model and the best bagging ensemble for each
classifier and over each MF and FE (found in section
4.2). It was found that bagging ensembles are
performing better than the single classifiers for KNN
(they have the highest accuracy), while for SVM and
MLP the single classifiers are performing better for
some FEs and MFs. For KNN, the difference of
accuracies between bagging ensembles is smaller
than the difference of accuracies between single
models (e.g., for MF 40×, the KNN single model has
an accuracy of 83.65% and 64.74% for DenseNet201
and MobileNetV2 respectively, while the bagging
KNN ensemble achieves an accuracy of 83.21% and
80.66% for DenseNet201 and MobileNetV2
respectively), which means that the KNN bagging
ensembles are more stable than the KNN single
models.
In order to identify which model is the best, the
models of the best SK cluster of each MF and FE were
ranked by using the Borda Count voting system. By
comparing the number of occurrences of bagging
ensembles and single techniques in the Borda Count
rankings results, it was found that single classifiers
are the most frequent for MLP (8 from 12) and SVM
Figure 5: Accuracy values of the bagging ensembles using KNN Classifier over each MF and FE.
Table 2: Performance values of single MLP classifiers and the best MLP bagging ensembles.
Single classifiers
(%)
Best Bagging ensembles
(%)
MF
FE
Accuracy Precision Sensitivity F1-score Accuracy Precision Sensitivity F1-score
40
×
DENSE
93.28 92.21 94.49 93.33 93.36 92.60 94.79 93.66
INC
92.92 91.30 94.90 93.05 93.87 93.73 94.53 94.11
MOB
91.06 88.94 93.82 91.30 89.93 88.01 93.41 90.61
100
×
DENSE
92.28 91.90 92.80 92.33 92.29 91.10 93.50 92.28
INC
90.22 88.85 92.25 90.43 90.97 88.09 94.51 91.17
MOB
91.16 89.20 93.67 91.38 88.33 84.92 92.90 88.70
200
×
DENSE
94.84 94.96 94.18 95.78 93.98 92.88 95.49 94.15
INC
91.76 91.85 91.87 91.94 90.68 90.74 91.01 90.82
MOB
91.80 90.70 93.24 91.92 89.18 87.04 92.66 89.72
400
×
DENSE
83.97 87.09 83.78 93.08 90.53 90.74 90.65 90.61
INC
90.28 90.49 88.90 92.24 89.64 88.25 91.60 89.90
MOB
89.77 87.70 92.54 90.05 88.94 85.61 93.62 89.41
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images
295

Table 3: Performance values of single SVM classifiers and the best SVM bagging ensembles.
Single classifiers (%) Best Bagging ensembles (%)
MF
FE
Accuracy Precision Sensitivity F1-score Accuracy Precision Sensitivity F1-score
40×
DENSE
92.52 90.98 94.36 92.62 91.61 90.73 93.41 92.02
INC
92.63 91.23 94.29 92.74 93.14 92.43 94.52 93.45
MOB
90.18 87.72 93.59 90.52 89.20 86.38 94.14 90.06
100×
DENSE
91.68 90.49 93.23 91.81 91.25 89.36 93.39 91.32
INC
90.15 87.85 93.27 90.46 90.97 87.74 95.05 91.22
MOB
90.71 87.75 94.66 91.06 87.57 83.70 92.77 87.98
200×
DENSE
93.60 92.45 94.97 93.68 93.91 93.12 95.08 94.07
INC
89.75 86.86 93.66 90.13 90.04 88.43 92.56 90.43
MOB
90.97 88.45 94.33 91.26 88.60 86.43 92.24 89.20
400×
DENSE
91.23 89.64 93.36 91.42 89.96 88.81 91.92 90.27
INC
88.79 86.30 92.40 89.21 90.63 87.13 95.07 90.91
MOB
89.16 86.29 93.21 89.59 87.57 83.70 92.77 87.98
Table 4: Performance values of single KNN classifiers and the best KNN bagging ensembles.
Single classifiers (%) Best Bagging ensembles (%)
MF
FE
Accuracy Precision Sensitivity F1-score Accuracy Precision Sensitivity F1-score
40×
DENSE
83.65 76.28 97.66 85.64 83.21 76.29 98.32 85.89
INC
82.66 79.05 88.98 83.68 82.99 78.63 92.43 84.96
MOB
64.74 58.73 99.42 73.82 80.66 73.50 98.31 84.09
100×
DENSE
85.04 79.27 94.95 86.40 84.72 78.60 94.78 85.89
INC
78.22 72.44 91.25 80.75 79.03 72.99 91.67 81.16
MOB
69.94 62.74 98.32 76.58 82.92 75.13 97.74 84.90
200×
DENSE
83.42 76.62 96.32 85.33 86.38 80.10 97.61 87.95
INC
79.50 73.59 92.16 81.81 80.79 74.41 95.00 83.43
MOB
70.90 63.54 98.27 77.16 81.36 74.35 96.95 84.12
400×
DENSE
80.67 73.95 94.96 83.10 76.03 70.19 91.85 79.53
INC
77.34 71.66 90.57 80.00 79.92 74.02 93.26 82.51
MOB
71.13 66.10 86.77 75.00 82.85 75.18 97.19 84.75
(7 from 12), but for KNN, the bagging ensembles
outperformed the single classifiers since 10 bagging
ensembles from 12 were ranked first.
In summary, for KNN, the bagging ensembles
outperformed the single classifiers for the majority of
MFs and FEs. Contrarily, MLP and SVM single
classifiers generally performed better than the
bagging ensembles. The reason is that MLP and SVM
can be considered as strong learners, and their single
classifiers significantly perform well compared to the
single KNN classifier.
4.4 Is There any Bagging Ensemble
that Outperforms the Others for
Each MF and over all the MFs?
This section evaluates and compares the best bagging
ensembles of each hybrid architecture (FE and
classifier) over each MF and the best ensembles over
all the MFs. Figure 6 shows the results of SK test for
the best bagging ensembles of each hybrid
architecture (found in section 4.2) over the four MFs.
Different clusters were obtained: four clusters for MF
40×, 100× and 200×, and three clusters for MF 400×.
In addition to the classifiers used in the present paper,
the bagging ensembles with DT classifier and
different FEs developed in this paper (Nakach et al.
2022b) were also added to the comparison (they are
abbreviated using “EBGD”).
It was found that SVM and MLP with
DenseNet201 and InceptionV3 always form the hybrid
architecture of the best four bagging ensembles for the
different MFs but they don’t respect a specific order
and they use a different number of base learners. Three
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
296
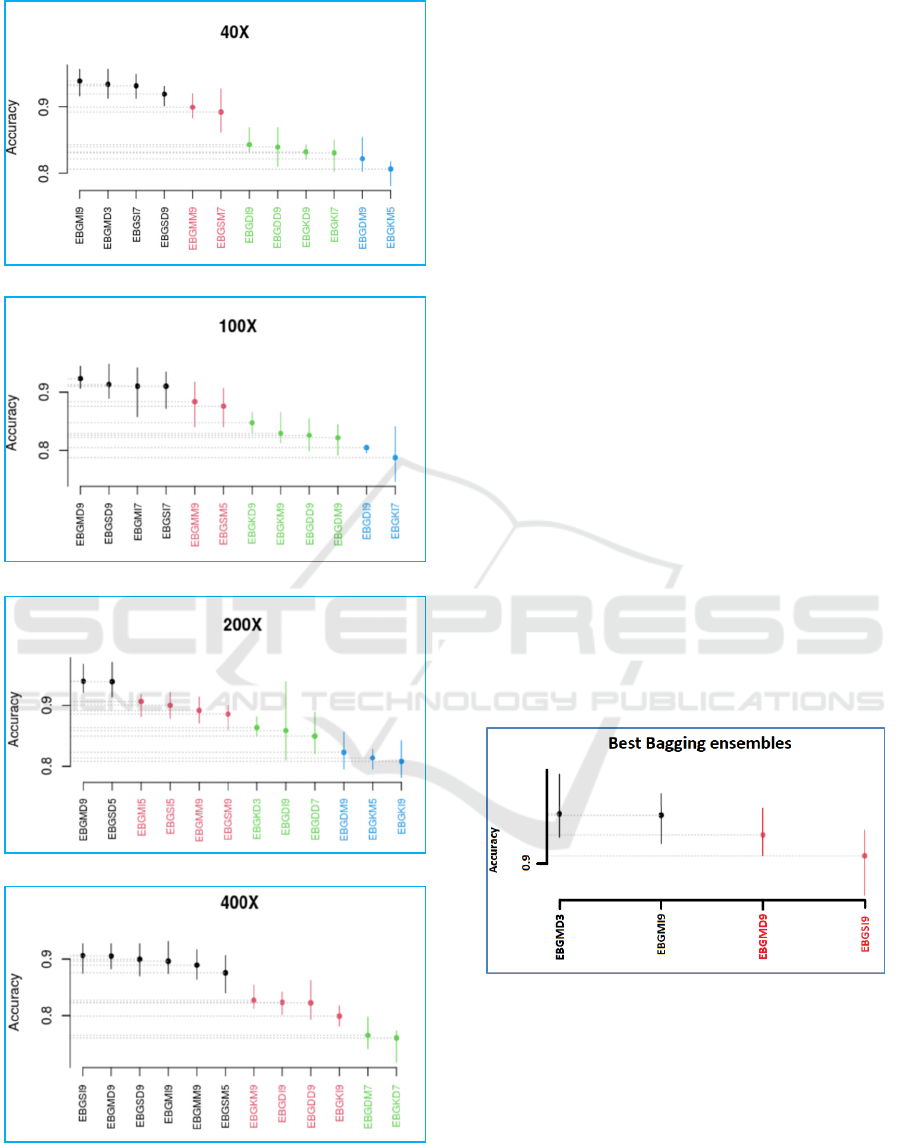
(a)
(b)
(c)
(d)
Figure 6: SK Results of the best bagging ensembles of each
classifier and FE over each MF: (a) 40×, (b) 100×, (c) 200×,
(d) 400×.
from the four first ranked ensembles were constructed
using MLP, and only the best bagging ensemble over
MF 400× was constructed with SVM. The majority of
those ensembles have 9 base learners (8 from 16) and
only 2 ensembles have 3 base learners. For
DenseNet201, MLP is ranked first for all the MFs, but
for InceptionV3 MLP is ranked first for MF 40× and
MF 200× while SVM is ranked first for MF 100× and
MF 400×.
The results show that the performance of the
different bagging ensembles depends on the
characteristic of the images and the bagging
ensembles that use MLP and SVM as base learners
outperform the bagging ensembles that use KNN and
DT as base learners. Figure 7 shows the results of SK
test for the best bagging ensembles over the four MFs.
Table 5 presents the ranking results of the best
bagging ensembles using the Borda count voting to
identify the best hybrid architecture regardless of the
MF.
To sum up, the bagging ensemble that uses MF 200×,
3 base learners and the hybrid architecture MLP with
DenseNet201 as FE was ranked first, the second-best
ensemble uses MF 40×, 9 base learners and the base
hybrid architecture MLP with InceptionV3 as FE, the
third best ensemble is constructed using MF 100×, 9
base learners and uses the base hybrid architecture
MLP with DenseNet201 as FE. The bagging
ensemble designed with MF 400×, 9 base learners and
whose base hybrid architecture is SVM with
InceptionV3 as FE was ranked fourth.
Figure 7: SK test Results of the best bagging ensembles.
In addition, the results obtained in the experimental
studies in the literature using the BreakHis dataset for
binary classification of the four MFs are presented for
comparison purposes in Table 6. It should be taken
into account that different training and test sample
numbers were used in these studies. The proposed
bagging ensembles can produce considerably higher
accuracies compared to the state-of-the-art model,
they achieve better performance than the candidate
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images
297

Table 5: Best bagging ensembles over all the MFs.
Model MF Accuracy (%) F1-score (%) Precision (%) Sensitivity (%) Rank
EBGMD3 200×
93,98 94,15 92,88 95,49
1
EBGMI9 40×
93,87 94,11 93,73 94,53
2
EBGMD9
100×
92,29 92,28 91,10 93,50
3
EBGSI9 400×
90,63 90,91 87,13 95,07
4
Table 6: Accuracy comparisons between the best bagging ensembles for each MF and the other models on BreakHis.
Method 40×
(%)
100×
(%)
200×
(%)
400×
(%)
SIFT + Stacked ensemble [50] 88.5 89.2 88.4 83.6
CNNs + SVM [51] 87.7 88.9 90.1 85.1
CNN [52] 89.6 85.0 82.8 80.2
Hybrid CNN ensemble [39] 85.7 84.2 84.9 80.1
ResNet50 + bagging [38] 92.5 92.0 - -
InceptionV3+GBT [53] 88.9 90.1 89.1 87.5
InceptionV4+GBT [53] 90.4 92.3 93.7 90.1
ResNetV1152+GBT [53] 92.5 94.8 95.8 90.5
Variant of AlexNet [54] 95.0 91.5 91.8 95.0
Our best bagging ensembles 93,9 92,3 94.0 90,6
models, expect for: the best model of the study (Senan
et al.) for MF 40× and 400×, and the
ResNetV1152+GBT model of the study (Vo et al.
2019) for MF 100× and 200×.
5 CONCLUSIONS
In conclusion, this paper presented and discussed the
results of an empirical comparative study of bagging
ensembles for BC histopathological image
classification. This study used the bagging method
with different number of base learners (3, 5, 7 and 9)
and twelve hybrid architectures (three classifiers:
KNN, MLP and SVM with three DL techniques for
feature extraction: DenseNet201, MobileNetV2 and
InceptionV3) over the BreakHis dataset with its four
MFs (40×, 100×, 200× and 400×). The main findings
of this study are:
RQ1. Do Bagging Ensembles Perform Better than
Their base Learners?
Bagging ensembles outperform their base learners for
MLP and KNN, but for SVM, the base learners often
perform better than the bagging ensembles.
RQ2. How the Number of Base Learners Impacts
the Performances of Bagging Ensembles?
For the majority of MF and FE, the bagging
ensembles designed with KNN and MLP respects an
ascending order where the bagging ensemble of 9
base learners represents the best ensemble and the
ensemble of 3 base learners represents the worst one.
RQ3. Do Bagging Ensembles Perform Better than
Single Classifiers?
For KNN, the bagging ensembles outperform the
single KNN classifiers for the majority of MF and FE.
Contrarily, MLP and SVM single classifiers
generally perform better than the bagging ensembles.
RQ4. Is There Any Bagging Ensemble that
Outperforms the Others for Each MF And Over
all the MFs?
The best bagging ensemble is designed using MF
200×, MLP as a classifier and DenseNet 201 as FE
and it has 3 base learners.
In conclusion, the proposed bagging ensembles
achieved promising results that can outperform state-
of-the-art models. Hence, this paper suggests that
bagging with hybrid DL architectures should be
considered for BC image classification. In a future
work, in order to deepen this analysis, it is possible to
implement heterogeneous ensemble learning methods
to classify BC histopathological images and compare
their performance with the bagging ensembles
implemented in this study.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
298

REFERENCES
Abbasniya MR, Sheikholeslamzadeh SA, Nasiri H, Emami
S (2022) Classification of Breast Tumors Based on
Histopathology Images Using Deep Features and
Ensemble of Gradient Boosting Methods. Computers
and Electrical Engineering 103:108382.
https://doi.org/10.1016/j.compeleceng.2022.108382
Abdar M, Makarenkov V (2019) CWV-BANN-SVM
ensemble learning classifier for an accurate diagnosis
of breast cancer. Measurement 146:557–570.
https://doi.org/10.1016/j.measurement.2019.05.022
Abdar M, Zomorodi-Moghadam M, Zhou X, et al (2020) A
new nested ensemble technique for automated
diagnosis of breast cancer. Pattern Recognition Letters
132:123–131. https://doi.org/10.1016/j.patrec.2018.11.
004
Alaoui O, Zerouaoui H, Idri A (2022) Deep Stacked
Ensemble for Breast Cancer Diagnosis. pp 435–445
B N (2019) Image Data Pre-Processing for Neural
Networks. In: Medium. https://becominghuman.ai/
image-data-pre-processing-for-neural-networks-49828
9068258. Accessed 12 May 2021
Breiman L (1996) Bagging predictors. Mach Learn 24:123–
140. https://doi.org/10.1007/BF00058655
Clegg LX, Reichman ME, Miller BA, et al (2009) Impact
of socioeconomic status on cancer incidence and stage
at diagnosis: selected findings from the surveillance,
epidemiology, and end results: National Longitudinal
Mortality Study. Cancer Causes Control 20:417–435.
https://doi.org/10.1007/s10552-008-9256-0
din NM ud, Dar RA, Rasool M, Assad A (2022) Breast
cancer detection using deep learning: Datasets,
methods, and challenges ahead. Computers in Biology
and Medicine 149:106073. https://doi.org/10.
1016/j.compbiomed.2022.106073
Emerson P (2013) The original Borda count and partial
voting. Soc Choice Welf 40:353–358. https://
doi.org/10.1007/s00355-011-0603-9
Guo Y, Hui D, Song F, et al (2018) Breast Cancer Histology
Image Classification Based on Deep Neural Networks.
pp 827–836
Gupta V, Bhavsar A (2017) Breast Cancer
Histopathological Image Classification: Is
Magnification Important? In: 2017 IEEE Conference
on Computer Vision and Pattern Recognition
Workshops (CVPRW). IEEE, Honolulu, HI, USA, pp
769–776
Güzel K, Bı̇ Lgı̇ N G (2020) Classification of Breast Cancer
Images Using Ensembles of Transfer Learning. Sakarya
University Journal of Science. https://doi.org/
10.16984/saufenbilder.720693
Hameed Z, Zahia S, Garcia-Zapirain B, et al (2020) Breast
Cancer Histopathology Image Classification Using an
Ensemble of Deep Learning Models. Sensors 20:4373.
https://doi.org/10.3390/s20164373
Hastie T, Tibshirani R, Friedman J (2009) Ensemble
Learning. In: Hastie T, Tibshirani R, Friedman J (eds)
The Elements of Statistical Learning: Data Mining,
Inference, and Prediction. Springer, New York, NY, pp
605–624
Jelihovschi E, Faria JC, Allaman IB (2014) ScottKnott: A
Package for Performing the Scott-Knott Clustering
Algorithm in R. Tend Mat Apl Comput 15:003.
https://doi.org/10.5540/tema.2014.015.01.0003
Kassani SH, Kassani PH, Wesolowski MJ, et al (2019)
Classification of Histopathological Biopsy Images
Using Ensemble of Deep Learning Networks.
arXiv:190911870 [cs, eess]
Kumar V, Abbas AK, Aster JC (2017) Robbins Basic
Pathology E-Book. Elsevier Health Sciences
Lbachir IA, Daoudi I, Tallal S (2021) Automatic computer-
aided diagnosis system for mass detection and
classification in mammography. Multimed Tools Appl
80:9493–9525. https://doi.org/10.1007/s11042-020-
09991-3
Nakach F-Z, Zerouaoui H, Idri A (2022a) Deep Hybrid
AdaBoost Ensembles for Histopathological Breast
Cancer Classification. In: Rocha A, Adeli H, Dzemyda
G, Moreira F (eds) Information Systems and
Technologies. Springer International Publishing,
Cham, pp 446–455
Nakach F-Z, Zerouaoui H, Idri A (2022b) Random Forest
Based Deep Hybrid Architecture for Histopathological
Breast Cancer Images Classification. pp 3–18
Nascimento DSC, Canuto AMP, Silva LMM, Coelho ALV
(2011) Combining different ways to generate diversity
in bagging models: An evolutionary approach. In: The
2011 International Joint Conference on Neural
Networks. pp 2235–2242
Naveen, Sharma RK, Ramachandran Nair A (2019)
Efficient Breast Cancer Prediction Using Ensemble
Machine Learning Models. In: 2019 4th International
Conference on Recent Trends on Electronics,
Information, Communication & Technology
(RTEICT). IEEE, Bangalore, India, pp 100–104
Nemade V, Pathak S, Dubey A, Barhate D (2022) A Review
and Computational Analysis of Breast Cancer Using
Different Machine Learning Techniques. https://doi.
org/10.46338/ijetae0322_13
Opitz D, Maclin R (1999) Popular Ensemble Methods: An
Empirical Study. jair 11:169–198. https://doi.org/
10.1613/jair.614
Otoom AF, Abdallah EE, Hammad M (2015) Breast Cancer
Classification: Comparative Performance Analysis of
Image Shape-Based Features and Microarray Gene
Expression Data. IJBSBT 7:37–46. https://doi.org/
10.14257/ijbsbt.2015.7.2.04
Polikar R (2012) Ensemble Learning. In: Zhang C, Ma Y
(eds) Ensemble Machine Learning: Methods and
Applications. Springer US, Boston, MA, pp 1–34
Ponnaganti ND, Anitha R (2022) A Novel Ensemble
Bagging Classification Method for Breast Cancer
Classification Using Machine Learning Techniques. TS
39:229–237. https://doi.org/10.18280/ts.390123
Saxena S, Gyanchandani M (2020) Machine Learning
Methods for Computer-Aided Breast Cancer Diagnosis
Using Histopathology: A Narrative Review. Journal of
Deep Hybrid Bagging Ensembles for Classifying Histopathological Breast Cancer Images
299

Medical Imaging and Radiation Sciences 51:182–193.
https://doi.org/10.1016/j.jmir.2019.11.001
Senan EM, Alsaade FW, Ahmed MI Classification of
Histopathological Images for Early Detection of Breast
Cancer Using Deep Learning. 24:7
Sharma S, Deshpande S (2021) Breast Cancer
Classification Using Machine Learning Algorithms. In:
Joshi A, Khosravy M, Gupta N (eds) Machine Learning
for Predictive Analysis. Springer, Singapore, pp 571–
578
Shen S, Sadoughi M, Li M, et al (2020) Deep convolutional
neural networks with ensemble learning and transfer
learning for capacity estimation of lithium-ion batteries.
Applied Energy 260:114296. https://doi.org/10.1016/
j.apenergy.2019.114296
Shorten C, Khoshgoftaar TM (2019) A survey on Image
Data Augmentation for Deep Learning. Journal of Big
Data 6:60. https://doi.org/10.1186/s40537-019-0197-0
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2016) A
Dataset for Breast Cancer Histopathological Image
Classification. IEEE Transactions on Biomedical
Engineering 63:1455–1462. https://doi.org/10.1109/
TBME.2015.2496264
Sung H, Ferlay J, Siegel RL, et al (2021) Global Cancer
Statistics 2020: GLOBOCAN Estimates of Incidence
and Mortality Worldwide for 36 Cancers in 185
Countries. CA: A Cancer Journal for Clinicians
71:209–249. https://doi.org/10.3322/caac.21660
Tuv E (2006) Ensemble Learning. In: Guyon I, Nikravesh
M, Gunn S, Zadeh LA (eds) Feature Extraction:
Foundations and Applications. Springer, Berlin,
Heidelberg, pp 187–204
Vo DM, Nguyen N-Q, Lee S-W (2019) Classification of
breast cancer histology images using incremental
boosting convolution networks. Information Sciences
482:123–138. https://doi.org/10.1016/j.ins.2018.12.089
Wang H, Zheng B, Yoon SW, Ko HS (2018) A support
vector machine-based ensemble algorithm for breast
cancer diagnosis. European Journal of Operational
Research 267:687–699. https://doi.org/10.1016/
j.ejor.2017.12.001
Wang J, Zhu T, Liang S, et al (2020) Binary and Multiclass
Classification of Histopathological Images Using
Machine Learning Techniques. Journal of Medical
Imaging and Health Informatics 10:2252–2258.
https://doi.org/10.1166/jmihi.2020.3124
Wu J, Hicks C (2021) Breast Cancer Type Classification
Using Machine Learning. Journal of Personalized
Medicine 11:61. https://doi.org/10.3390/jpm11020061
Yadavendra, Chand S (2020) A comparative study of breast
cancer tumor classification by classical machine
learning methods and deep learning method. Machine
Vision and Applications 31:46. https://doi.org/10.
1007/s00138-020-01094-1
Yussof W (2013) Performing Contrast Limited Adaptive
Histogram Equalization Technique on Combined Color
Models for Underwater Image Enhancement
Zerouaoui H, Idri A (2021) Reviewing Machine Learning
and Image Processing Based Decision-Making Systems
for Breast Cancer Imaging. J Med Syst 45:8.
https://doi.org/10.1007/s10916-020-01689-1
Zerouaoui H, Idri A (2022) Deep hybrid architectures for
binary classification of medical breast cancer images.
Biomedical Signal Processing and Control 71:103226.
https://doi.org/10.1016/j.bspc.2021.103226
Zerouaoui H, Idri A, Nakach FZ, Hadri RE (2021) Breast
Fine Needle Cytological Classification Using Deep
Hybrid Architectures. In: Gervasi O, Murgante B,
Misra S, et al. (eds) Computational Science and Its
Applications – ICCSA 2021. Springer International
Publishing, Cham, pp 186–202
Zhang Y, Zhang B, Coenen F, Lu W (2013) Breast cancer
diagnosis from biopsy images with highly reliable
random subspace classifier ensembles. Machine Vision
and Applications 24:1405–1420. https://doi.org/10.
1007/s00138-012-0459-8
Zhu C, Song F, Wang Y, et al (2019) Breast cancer
histopathology image classification through assembling
multiple compact CNNs. BMC Med Inform Decis Mak
19:198. https://doi.org/10.1186/s12911-019-0913-x.
ICAART 2023 - 15th International Conference on Agents and Artificial Intelligence
300
