
vEEGNet: A New Deep Learning Model to Classify and Generate EEG
Alberto Zancanaro
1 a
, Italo F. Zoppis
2 b
, Sara L. Manzoni
2 c
and Giulia Cisotto
1,2 d
1
Department of Information Engineering, University of Padova, via Gradenigo 6/b, Padova, Italy
2
Department of Informatics, Systems, and Communications, University of Milano-Bicocca, viale Sarca 336, Milan, Italy
Keywords:
AI, Deep Learning, Variational Autoencoder, EEG, Machine Learning, Brain, Classification, Latent Space,
Inter-Subject Variability.
Abstract:
The classification of EEG during motor imagery (MI) represents a challenging task in neuro-rehabilitation. In
2016, a deep learning (DL) model called EEGNet (based on CNN) and its variants attracted much attention
for their ability to reach 80% accuracy in a 4-class MI classification. However, they can poorly explain
their output decisions, preventing them from definitely solving questions related to inter-subject variability,
generalization, and optimal classification. In this paper, we propose vEEGNet, a new model based on EEGNet,
whose objective is now two-fold: it is used to classify MI, but also to reconstruct (and eventually generate)
EEG signals. The work is still preliminary, but we are able to show that vEEGNet is able to classify 4 types
of MI with performances at the state of the art, and, more interestingly, we found out that the reconstructed
signals are consistent with the so-called motor-related cortical potentials, very specific and well-known motor-
related EEG patterns. Thus, jointly training vEEGNet to both classify and reconstruct EEG might lead it,
in the future, to decrease the inter-subject performance variability, and also to generate new EEG samples to
augment small datasets to improve classification, with a consequent strong impact on neuro-rehabilitation.
1 INTRODUCTION
Electroencephalography (EEG)-based classification
represents a challenging and critical problem in many
applications, e.g., neuroscience and brain–computer
interface (BCI) to support the diagnosis of move-
ment disorders and motor rehabilitation (Cisotto et al.,
2022). Particularly, besides promising achievements
in supporting disabled individuals, neurorobotics and
BCI systems (Beraldo et al., 2022) are still poorly
performing in many tasks, e.g., motor imagery (MI)
classification. There exist several machine learning
(ML) and deep learning (DL) models to classify EEG
of imagined movements: filter-bank common spa-
tial pattern (FBCSP) (Kai Keng Ang et al., 2008) is
the standard ML model, very common in BCI ap-
plications where it is used also in real-time. More
recently, convolutional neural networks (CNN) have
gained a lot of attention as architectures particularly
good in classifying EEG. In 2016, EEGNet, an ar-
chitecture made of 2 blocks, each one composed of
a
https://orcid.org/0000-0002-5276-7030
b
https://orcid.org/0000-0001-7312-7123
c
https://orcid.org/0000-0002-6406-536X
d
https://orcid.org/0000-0002-9554-9367
2 convolutional layers and a fully-connected layer,
was published by (Lawhern et al., 2016). Given its
success in classifying EEG in different classes of
movements (both executed and imagined), a num-
ber of variants were presented, including Temporary
Constrained Sparse Group Lasso enhanced EEGNet
(TSGL-EEGNet) (Deng et al., 2021), Multibranch
Shallow CNN (MBShallow ConvNet) (Altuwaijri and
Muhammad, 2022), MI-EEGNet (Riyad et al., 2021),
Quantized EEGNet (Q-EEGNet) (Schneider et al.,
2020), DynamicNet (Zancanaro et al., 2021), and
other general-purpose CNN models, namely Channel-
wise CNN (CW-CNN) (Sakhavi et al., 2018), Densely
Feature Fusion CNN (DFFN) (Li et al., 2019a),
and the Monolithic Network (Olivas and Chacon,
2018). They differ from each other by a more (e.g.,
MI-EEGNet) or less (e.g., EEGNet) invasive pre-
processing of the EEG signal, by their architectures
with single or multiple EEGNet units combined to
extract one or a few sets of artificial features (e.g.,
TSGL-EEGNet and MBShallow ConvNet), and by
their feasibility in running on portable devices (e.g.,
Q-EEGNet).
They achieve accuracies in the range of 70% to
80% in a 4-class MI classification. However, they
Zancanaro, A., Zoppis, I., Manzoni, S. and Cisotto, G.
vEEGNet: A New Deep Learning Model to Classify and Generate EEG.
DOI: 10.5220/0011990800003476
In Proceedings of the 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2023), pages 245-252
ISBN: 978-989-758-645-3; ISSN: 2184-4984
Copyright
c
2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
245

cannot, or poorly, relate their classification decisions
with well-known EEG patterns or biomarkers.
In this paper, we aim to propose our own DL
model, named as vEEGNet, whose objective is two-
fold: on one side, the model is used to classify EEG
signals obtained during the participant’s MI of dif-
ferent body segments (i.e., one hand, the feet, or the
tongue); on the other side, the model is enriched by
a generative module that is able to reconstruct some
specific EEG components, strongly related to MI.
vEEGNet consists of two learning modules, i.e., an
unsupervised representation learning module, and a
supervised module. The first one is formed by a vari-
ational auto-encoder (VAE) (Kingma and Welling,
2013; Zancanaro et al., 2022; Li et al., 2019b), while
the second is implemented using a feed-forward neu-
ral network (FFNN). In the VAE, we exploit EEGNet
as an encoder (and, conversely, its mirrored version as
a decoder) to extract a compact and highly informa-
tive representation of the EEG. The encoder extracts
a compact and latent representation of the EEG that is
later used by the FFNN to classify the EEG into four
different classes of movement. At the same time, that
representation made it possible to generate new syn-
thetic EEG samples. To take advantage of this com-
bined approach, vEEGNet was trained by minimizing
a joint loss function given by the sum of the VAE loss
and the classifier loss.
To assess the performance of vEEGNet as classi-
fier, we tested it on the public dataset 2a from the BCI
competition IV (containing EEG during four types
of imagined movements) and compared the results
with other models based on EEGNet that were pre-
viously employed to classify the same dataset. We
show that vEEGNet reaches comparable classification
accuracies and Cohen’s κ score as the state of the
art (approximately ranging between 70% and 80%).
Then, we investigated its ability to decode a multi-
channel EEG from its latent representation and we
might speculate that our model is able to reconstruct
a particular low-frequency well-known component of
the EEG that is related to any executed or imagined
movement, i.e., the motor related cortical potential
(MRCP). However, this contribution is still prelimi-
nary and, as such, a number of limitations and open
challenges are also discussed, and will need further
investigations. Nevertheless, this paper represents a
promising way to shed more light on the ability of DL
models to solve very complex tasks, such as recog-
nizing different imagined movements from an EEG,
providing a link to common neurophysiological pat-
terns that the model might be able to identify and also
generate. Furthermore, this paper can contribute to
the research question of how to eventually augment
EEG datasets, that typically suffer from limited sizes,
preventing DL models to reach satisfactory levels of
robustness and generalization.
The rest of this paper is organized as follows: Sec-
tion 2 describes the VAE theory and introduces the
vEEGNet model. Section 3 presents the classification
results with respect to other CNN or EEGNet-based
models, and discusses the reconstruction and gener-
ative potentialities of vEEGNet. Finally, section 4
concludes the paper and paves the way toward new
promising future directions.
2 MATERIALS AND METHODS
2.1 Variational Autoencoder
VAE is an effective encoding-decoding DL approach
that provides a structured latent space to be used
for random sampling and interpolation (Kingma and
Welling, 2013). These properties have led to ef-
ficient implementations of VAEs for several unsu-
pervised and semi-supervised learning problems (see
e.g., (Hinton and Salakhutdinov, 2006; Li et al.,
2019b; Zancanaro et al., 2022)). In probabilistic
terms, a VAE is able to learn a variational (approx-
imate posterior) distribution q
φ
(z|x) of latent vari-
ables z, given the observations x, as well as a gen-
erative model p
θ
(x|z) (Blei et al., 2017). This task is
obtained using an encoder-decoder pair of deep net-
works parametrized by φ and θ, respectively. The
training consists of the minimization (w.r.t. param-
eters φ and θ) of the VAE loss, L
VAE
. Typically,
the VAE loss is expressed in terms of evidence lower
bound (ELBO) for the (evidence) probability p(x),
namely L(θ,φ;x): L
VAE
= −L(θ, φ;x), provided that
L(θ,φ;x) = E
q
φ
(z|x)
log
p
θ
(x,z)
q
φ
(z|x))
(1)
Thus, for the VAE training, minimizing L
VAE
means
maximizing the ELBO for p(x). The gap between
p(x) and L(θ,φ; x) can be best expressed by consid-
ering the Kullback-Leibler divergence (K L) between
the variational q
φ
(z|x) and posterior p
θ
(x|z) distribu-
tions, which turns to be
K L[q
φ
(z|x)||p
θ
(x|z)] = −L(θ, φ;x)+ p(x) (2)
Since K L[q
φ
(z|x)||p
θ
(x|z)] ≥ 0, one arrives at the
lower bound L(θ,φ;x) ≤ p(x). Similarly, ELBO can
be also formulated as
L(θ,φ;x) = E
q
(p
θ
(x|z)) − K L[q
φ
(z|x)||p(z)] (3)
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
246

In this way, the second term K L[q
φ
(z|x)||p(z)] acts
as a regularizer, thus penalizing those surrogate dis-
tributions, q
φ
(z|x), too far away from the predefined
p(z).
2.2 vEEGNet
In this work, we devised a new combined model based
on a VAE (Kingma and Welling, 2013; Li et al.,
2019b) and our previous implementation of EEG-
Net (Zancanaro et al., 2021), as represented in Fig. 1.
Particularly, the model exploits EEGNet in the VAE,
for both encoding and decoding the EEG samples,
while an FFNN is used for the classification. As
a consequence, the model consists of two different
mechanisms, ruled by an unsupervised and a super-
vised learning, respectively, as further explained in
the following.
2.2.1 Unsupervised Mechanism
The unsupervised mechanism (i.e., the VAE) exploits
the EEGNet architecture to supply the latent distri-
bution q
φ
(z|x) as well as the posterior p
θ
(x|z). We
assumed isotropic Gaussian distribution for both the
prior p(z) and the approximate posterior, q
φ
(z|x), i.e.,
p(z) = N (0,I) (4)
q
φ
(z|x) = N (z;µ(x; φ),σ
2
(x;φ)I
I
I) (5)
where µ(x; φ) and σ(x; φ) are the functions imple-
mented by the vEEGNet encoder to encode the
mean and the (diagonal) covariance matrix of the
Gaussian distribution. With these assumptions,
K L[q
φ
(z|x)||p(z)] (the regularization term defined in
Section 2.1) can be directly expressed in the compact
analytical form (Kingma and Welling, 2013):
L
KL
= K L[q
φ
(z|x)||p(z)] =
1
2
d
∑
i=1
(σ
2
i
+ µ
2
i
− 1 − log(σ
2
i
))
(6)
where µ
i
and σ
2
i
are the predicted mean and variance
values of the corresponding i-th latent component
of z. The vEEGNet encoder implements a standard
EEGNet with its usual blocks, i.e., a temporal convo-
lution, a spatial convolution, and a separable convolu-
tion. Lastly, the output is flattened and passed through
a fully-connected layer. From the vEEGNet encoder’s
output (i.e., giving q
φ
(z|x)), we sample a vector, say
z
1
1
, and provide it as the input for the vEEGNet de-
coder that has the final aim to reconstruct the origi-
nal EEG signal. The vEEGNet decoder implements
1
Because this operation is not differentiable this is typ-
ically obtained with reparametrization by setting z
1
= µ +
σ · N(0,1
1
1).
a mirrored EEGNet structure using transposed convo-
lutions (in place of the standard convolution) and up-
sample layers (in place of the pooling layers). In both
the vEEGNet encoder and the decoder, batch normal-
ization and dropout layers were added to increase per-
formance and stability during training.
2.2.2 Supervised Mechanism
The supervised mechanism is given by an FFNN that
classifies the EEG into 4 different classes. The FFNN
consists of an input layer (128 neurons), followed by
one hidden layer (64 neurons) and one output layer (4
neurons) for the target. In vEEGNet, a second vector
z
2
= [µ
µ
µ,σ
σ
σ
2
] is obtained by concatenating the output
of the encoder, i.e. the parameters vectors ˜µ = µ(x; φ
φ
φ)
and
˜
σ = σ(x; φ
φ
φ). This new vector is fed into the clas-
sifier to output the predicted class ˜y. For the classifier,
we used the negative log-likelihood loss function de-
fined as:
L
cl f
= − log(
˜
y) · y (7)
where log(
˜
y) are the log probabilities of possible la-
bels related to input x, and y is a one hot encoded
vector of the true labels of input x.
Overall, vEEGNet aims to minimize the loss func-
tion L
Total
given by the sum of the VAE loss func-
tion and the classifier loss function (L
cl f
), as follows:
L
Total
= L
VAE
+ L
cl f
.
3 RESULTS AND DISCUSSION
3.1 Dataset and vEEGNet
Implementation
To test the reliability of vEEGNet as a model for
EEG-based MI, we used it to classify the 4 differ-
ent MI tasks included in the public dataset 2a of the
IV BCI competition (Blankertz et al., 2007). The
latter includes 22-channel EEG recordings from 9
subjects repeatedly performing MI of either right or
left hand, feet or tongue. A set of 288 trials were
available for each subject for the training, and an-
other set of 288 trials for the test set for each sub-
ject. The EEG data have been previously filtered with
a 0.5 − 100Hz band-pass filter and a notch filter at
50 Hz. In line with other works (Riyad et al., 2021;
Lawhern et al., 2016) and our previous paper (Zan-
canaro et al., 2021), we down-sampled the EEG sig-
nals at 128Hz. Then, from each MI repetition, one
4s multi-channel EEG segment was extracted, thus
obtaining a 22 × 512 data matrix. We implemented
vEEGNet: A New Deep Learning Model to Classify and Generate EEG
247
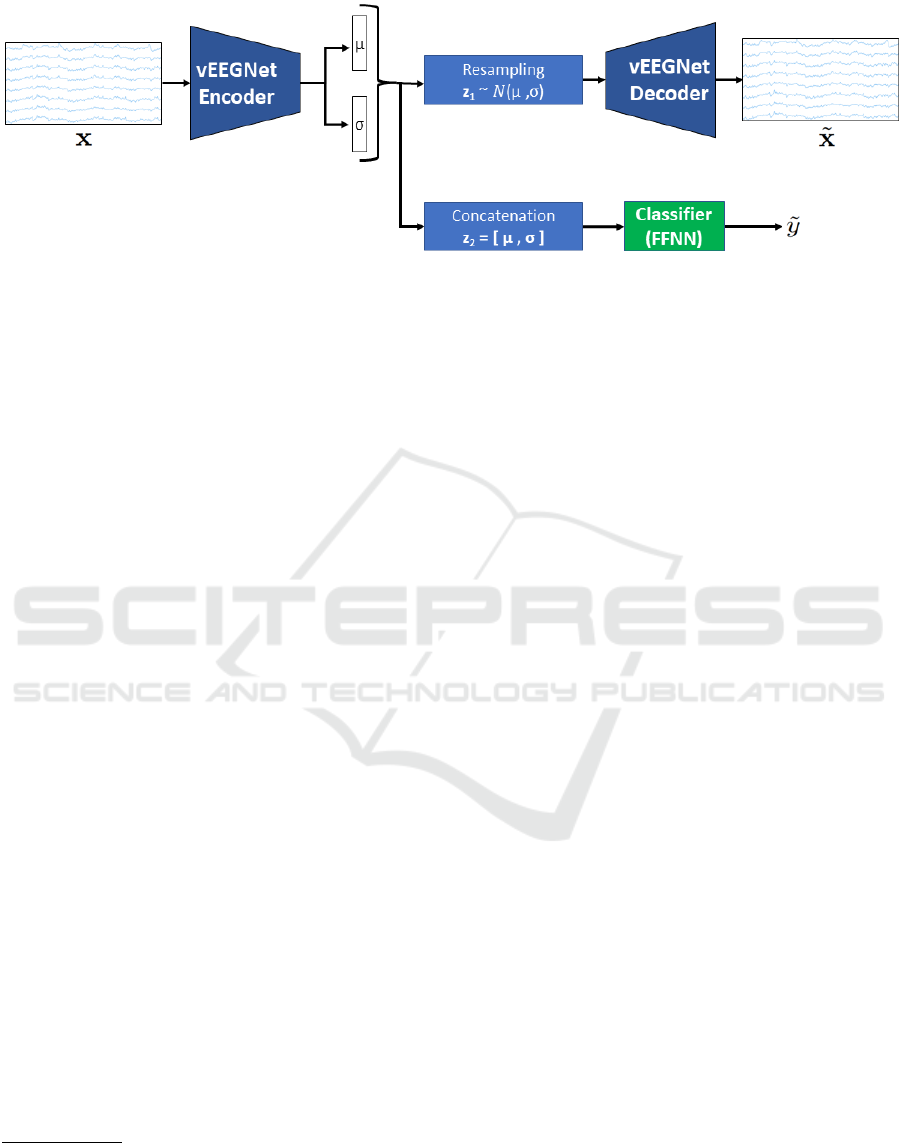
Figure 1: vEEGNet architecture.
vEEGNet in PyTorch
2
and we trained it using RTX
2070, 500 epochs, AdamW optimizer (Loshchilov
and Hutter, 2019), a learning rate of 0.001, and a
weight decay of 0.00001. The total number of train-
able parameters is 61476, with 52960 of them for the
implementation of the unsupervised mechanism and
the remaining 8516 for the supervised one. We em-
pirically chose d = 16 as the hidden space dimension.
In line with a common empirical approach (the in-
terested reader can refer to the TensorFlow Tutorial
3
,
we considered the first d/2 neurons as µ
µ
µ vector of the
means, and the remaining d/2 neurons account for
the variance σ
σ
σ
2
2
2
vector. Incidentally, we report that we
have tested the results for different values of d, specif-
ically, d = 2,4,8,16, 32,64,128, finding comparable
results.
3.2 vEEGNet as Classifier
vEEGNet was used to classify the MI class for every
subject in the dataset. Table 1 reports its classifica-
tion performance in terms of accuracy and Cohen’s
κ score with respect to other DL models, including
our previous optimized implementation of EEGNet
(DynamicNet (Zancanaro et al., 2021)) and general-
purpose CNN-based models (i.e., the CW-CNN, the
DFFN, and the Monolithic network). Performance are
reported for each individual subject as well as for the
grand-average (i.e., mean across all subjects).
We decided to include in the comparison only
those papers which reported the individual perfor-
mance for all subjects for the 4-class classification.
Thus, we excluded some previous works implement-
ing CNN- or EEGNet-based architectures that either
considered 2 classes or grand-average accuracy, only
(e.g., (Schirrmeister et al., 2017)). From Table 1,
2
The code is available on GitHub:
https://github.com/jesus-333/Variational-Autoencoder-
for-EEG-analysis
3
Available at https://www.tensorflow.org/tutorials/
generative/cvae
it can be noticed that those models which combine
multiple EEGNet units (e.g.. TSGL-EEGNet, MB-
Shallow ConvNet) can reach higher performance, in
the order of 80% (despite of the type of combina-
tion, i.e., in parallel or in series), while other mod-
els achieve accuracy values in the range 71%-78%. It
might be possible that this is due to the different fea-
tures that each specific architecture can extract, lead-
ing to better adaptability to each individual subject.
It is well-known that different subjects share similar
frequency bands to realize MI, but each of them can
have the strongest MI-related component at a slightly
different frequency (Magnuson and McNeil, 2021; Li
et al., 2018; Bressan et al., 2021). In turn, this might
be the reason why models built on a single choice of
frequency-domain features, i.e., including the origi-
nal EEGNet, are not able to generalize well. Also, it
is worth observing that most of the models, includ-
ing ours, apply very basic or no pre-processing at all.
MI-EEGNet is the only EEGNet-based model which
invasively pre-processes the input EEG with a nar-
row band 4-38 Hz filter and a 50 Hz notch, reach-
ing an accuracy value of 74.61% with very high vari-
ability across subjects (i.e., the standard deviation is
15.44%). At the individual subject level, from Ta-
ble 1, we found that there exists a large inter-subject
variability, as expected from the literature on EEG,
with standard deviation values in the range of 6.27%
to 15.44%. At the same time, it is not fully clear why
the classification accuracy for some specific subjects
(e.g., subject nn.3 and 7) is very high, despite the
model used, while for some others the classification
seems to be generally more difficult (e.g., for subject
nn.2 and 6). This requires further investigations in the
future to increase the adaptability and the generaliza-
tion ability of these kinds of DL architectures.
3.3 vEEGNet as Generator
Fig. 2 reports an example of reconstructed EEG sig-
nal from channel C3 during the imagination of the
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
248

Table 1: Comparison of vEEGNet with other DL models in terms of classification accuracy ([%]) and kappa score (when
available, its value is within brackets) in a four classes MI task. The first five columns refer to EEGNet-based models, while
the last three columns refer to general-purpose CNN models. AVG stands for average, STD for standard deviation.
vEEGNet
(d = 16)
EEGNet
(DynamicNet)
TSGL-
EEGNet
MI-EEGNet
MBShallow
ConvNet
CW-CNN DFFN
Monolithic
Network
1 78.13 (0.71) 81.88 85.41 (0.81) 83.68 (0.78) 82.58 (0.77) 86.11 (0.82) 83.2 83.13 (0.67)
2 61.81 (0.49) 60.97 70.67 (0.61) 49.65 (0.33) 70.01 (0.6) 60.76 (0.48) 65.69 65.45 (0.35)
3 84.72 (0.8) 88.54 95.24 (0.94) 89.24 (0.86) 93.79 (0.92) 86.81 (0.82) 90.29 80.29 (0.65)
4 65.28 (0.54) 70.63 80.26 (0.74) 68.06 (0.57) 82.6 (0.77) 67.36 (0.57) 69.42 81.6 (0.62)
5 70.49 (0.61) 68.45 70.29 (0.6) 64.93 (0.53) 77.81 (0.7) 62.5 (0.5) 61.65 76.7 (0.58)
6 60.42 (0.47) 61.46 68.37 (0.58) 56.25 (0.42) 64.79 (0.53) 45.14 (0.27) 60.74 71.12 (0.45)
7 79.86 (0.73) 82.08 90.97 (0.88) 94.1 (0.92) 88.02 (0.84) 90.63 (0.88) 85.18 84 (0.69)
8 79.17 (0.72) 82.15 86.35 (0.82) 82.64 (0.77) 86.91 (0.83) 81.25 (0.75) 84.21 82.66 (0.7)
9 67.71 (0.57) 66.25 83.64 (0.79) 82.99 (0.77) 83.38 (0.78) 77.08 (0.69) 85.48 80.74 (0.64)
AVG 71.95 (0.63) 73.60 81.34 (0.75) 74.61 (0.66) 81.15 (0.75) 73.07 (0.64) 76.44 78.1 (0.59)
STD 8.78 (0.12) 10.20 9.61 (0.13) 15.44 (0.21) 9.03 (0.12) 15.11 (0.2) 11.65 6.27 (0.12)
Figure 2: An example of reconstructed EEG (channel C3).
right-hand movement. At a first sight, the reconstruc-
tion seems not to be successful and poorly consistent
with the original signal. However, we might recog-
nize in the reconstructed signal a specific EEG com-
ponent that typically appears, following a precise tim-
ing, when a movement is executed or imagined, the
so-called MRCP. MRCPs are low-frequency compo-
nents (typically in the δ or θ bands, i.e., in the range
0.5-4 Hz) that are characterized by a sequence of pos-
itive and negative peaks after the ”GO” cue (i.e., the
time zero in our case) (Magnuson and McNeil, 2021).
Fig. 3 shows four different reconstructed EEG
channels, namely C3, C4, Cz, and the average of FC3
and FC4, selected based on their relevance to the MI
tasks. To be specific, in line with well-known lit-
erature (Lazurenko et al., 2018), the most relevant
electrodes where to retrieve information related to the
hand movement are the controlateral central sensors
C3 and C4, for the right and the left-hand move-
ments, respectively, while for the legs is Cz, and for
Figure 3: Reconstructed channels C3, C4, Cz, and average
FC3 and FC4.
the tongue are the frontal sensors F3 and F4 (with a
prevalence of F3). In our dataset, F3 and F4 were
not available, then we considered the nearest available
sensors which were FC3 and FC4 (as in the Interna-
tional 10-20 System for EEG electrode placement). If
Fig. 3 is compared with the consolidated literature on
MRCP during motor execution and imagery (Magnu-
son and McNeil, 2021; Li et al., 2018; Bressan et al.,
2021), we might recognize a very similar pattern: a
positive peak occurs right after the ”GO” cue, then a
negative peak follows (before 1 s), and finally a re-
bound is observed. The entire waveform almost ex-
pires (i.e., returns to baseline) within approximately
2 s after the cue. Here, we could observe a pattern
that is very consistent with the expected one. There-
fore, we can conclude that vEEGNet is extracting a
compact representation of a multi-channel EEG that
represents its lower frequency component during the
MI. This allows the model to obtain satisfactory accu-
vEEGNet: A New Deep Learning Model to Classify and Generate EEG
249

racy values in the classification of 4 different MI tasks
and to extract an MRCP pattern. However, this specu-
lation needs to be confirmed with further analysis and
investigations. Also, in the future, it be might worth
providing further explanations, in line with (Zoppis
et al., 2020; Scapin et al., 2022), of the mechanisms
that the DL models process the EEG signals, and how
to drive the architecture to reconstruct not only the
slower components of the signal (e.g., the MRCPs)
but also the faster ones (e.g., the µ and β components
ranging between 8 and 30 Hz) (Pfurtscheller et al.,
2006).
4 CONCLUSIONS
In this work, we tackled the challenging problem of
the multi-class classification of different MI tasks us-
ing EEG. Several ML and DL models have been pro-
posed to solve this complex problem. Among oth-
ers, EEGNet by (Lawhern et al., 2016) and its several
variants gained a lot of attention in the last few years,
since 2016. However, these models typically pro-
vide medium to high accuracy values (between 70%
and 80% approximately), but can poorly explain how
they decide on the classification output. Therefore,
in this work, we proposed a new DL model, namely
vEEGNet, whose objective is two-fold: the model
is used to classify EEG signals during participants’
MI (i.e., of a hand, the feet, or the tongue); at the
same time, it is enriched by a module that is able to
reconstruct the EEG. In vEEGNet, we employed an
EEGNet to encode a multi-channel EEG dataset, and
to extract a latent representation in e.g., 16 dimen-
sions. Then, a mirrored version of EEGNet is used to
decode such compact representation into a new syn-
thetically generated multi-channel EEG. In parallel,
a FFNN takes in input newly generated EEG sam-
ples from the latent representation and uses them to
recognize one out of four different imagined move-
ments. We show that vEEGNet is able to classify the
EEG with performances that are comparable with the
state of the art. Interestingly, we also found out that
the reconstructed signals resemble some specific, and
well-known, EEG components that are strongly re-
lated to MI, the MRCPs. Thus, this paper presents
a new architecture that has the potentiality to both
classify EEG during MI as well as provide a link
between neurophysiology and the model’s classifica-
tion decisions. Although vEEGNet, in its current im-
plementation, cannot significantly outperform other
models, it is worth highlighting that it was built on
top of the standard EEGNet (implemented in our Dy-
namicNet framework (Zancanaro et al., 2021)) and it
can achieve its reference state-of-the-art performance,
i.e., the fairest comparison being with EEGNet itself
which - in fact - reached a very close average accuracy
value, slightly exceeding 70%, across the subjects.
Thus, at present, we could not obtain a clear advan-
tage in terms of classification accuracy in having also
trained the model on the reconstruction term. This
is one of the limitations of this contribution. There
are also other aspects that will deserve further inves-
tigation. Particularly, it might be worth exploring,
at least, two different directions: on one side, opti-
mizing the overall loss function by better balancing
its two main contributions (i.e., the VAE loss func-
tion and the classification loss function) might lead
to a performance improvement. On the other hand,
another DL model, which can reach higher accura-
cies in its basic architecture compared to the standard
EEGNet (e.g., MBShallow ConvNet), might be used
to implement the encoder of vEEGNet to test if per-
formances increase in our more general-purpose ar-
chitecture. Another way to improve this work, and ex-
plain the vEEGNet model performance in both clas-
sification and reconstruction, as well as their mutual
relationship, is to study the different behavior of the
model in response to modifications of the input (as
in some explainability studies where ablation, permu-
tation or other kinds of perturbations have been ap-
plied to the EEG input (Manjunatha and Esfahani,
2021)), towards a more transparent and explainable
DL approach. Finally, modifications of the architec-
ture could be adopted to extract different features and
also reconstruct faster components that can be rele-
vant to the MI task, e.g., the α and β frequencies in
the range 8 to 30 Hz (as well-established by previ-
ous literature (Pfurtscheller et al., 2006)). Besides,
vEEGNet could be used to deepen into the problem of
the inter-subject variability that typically prevents DL
models to be easily generalized from subject to sub-
ject (and even experimental session to session of the
same subject). This might be of such an impact in the
field of, e.g., BCI, where the system needs to seam-
lessly interact with patients and healthy na
¨
ıve users.
Finally, future investigations of the potentialities of
vEEGNet as a generative model for EEG can be ad-
dressed to cope with the common lack of large EEG
datasets that make it difficult for DL models to im-
prove their performance and better generalize.
ACKNOWLEDGEMENTS
AZ is supported by PON 2014-2020 action IV.4
funded by the Italian Ministry of University and Re-
search at the University of Padova (Padova, Italy).
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
250

GC is supported by PON Initiative 2014-2020 action
IV.6 funded by the Italian Ministry of University and
Research at the University of Milan-Bicocca (Milan,
Italy).
REFERENCES
Altuwaijri, G. A. and Muhammad, G. (2022). A multi-
branch of convolutional neural network models for
electroencephalogram-based motor imagery classifi-
cation. Biosensors, 12(1).
Beraldo, G., Tonin, L., Mill
´
an, J. d. R., and Menegatti, E.
(2022). Shared intelligence for robot teleoperation via
BMI. IEEE Transactions on Human-Machine Sys-
tems.
Blankertz, B., Dornhege, G., Krauledat, M., M
¨
uller, K.-R.,
and Curio, G. (2007). The non-invasive Berlin Brain-
Computer Interface: Fast acquisition of effective per-
formance in untrained subjects. NeuroImage, 37:539–
50.
Blei, D. M., Kucukelbir, A., and McAuliffe, J. D.
(2017). Variational inference: A review for statisti-
cians. Journal of the American statistical Association,
112(518):859–877.
Bressan, G., Cisotto, G., M
¨
uller-Putz, G. R., and Wriess-
negger, S. C. (2021). Deep learning-based classifica-
tion of fine hand movements from low frequency eeg.
Future Internet, 13(5):103.
Cisotto, G., Capuzzo, M., Guglielmi, A. V., and Zanella,
A. (2022). Feature stability and setup minimiza-
tion for EEG-EMG-enabled monitoring systems.
EURASIP Journal on Advances in Signal Processing,
2022(1):103.
Deng, X., Zhang, B., Yu, N., Liu, K., and Sun, K.
(2021). Advanced TSGL-EEGNet for motor imagery
EEG-based Brain-Computer Interfaces. IEEE Access,
9:25118–25130.
Hinton, G. E. and Salakhutdinov, R. R. (2006). Reducing
the dimensionality of data with neural networks. sci-
ence, 313(5786):504–507.
Kai Keng Ang, Zheng Yang Chin, Haihong Zhang, and
Cuntai Guan (2008). Filter bank common spatial
pattern (FBCSP) in Brain-Computer Interface. In
2008 IEEE Int. Joint Conf. on Neural Networks (IEEE
World Congress on Computational Intelligence).
Kingma, D. P. and Welling, M. (2013). Auto-encoding vari-
ational bayes. arXiv preprint arXiv:1312.6114.
Lawhern, V., Solon, A., Waytowich, N., Gordon, S., Hung,
C., and Lance, B. (2016). EEGNet: A compact convo-
lutional network for EEG-based Brain-Computer In-
terfaces. Journal of Neural Engineering, 15.
Lazurenko, D., Kiroy, V., Aslanyan, E., Shepelev, I.,
Bakhtin, O., and Minyaeva, N. (2018). Electrographic
properties of movement-related potentials. Neu-
roscience and Behavioral Physiology, 48(9):1078–
1087.
Li, D., Wang, J., Xu, J., and Fang, X. (2019a). Densely
feature fusion based on convolutional neural networks
for motor imagery EEG classification. IEEE Access,
7:132720–132730.
Li, H., Huang, G., Lin, Q., Zhao, J.-L., Lo, W.-L. A., Mao,
Y.-R., Chen, L., Zhang, Z.-G., Huang, D.-F., and Li,
L. (2018). Combining movement-related cortical po-
tentials and event-related desynchronization to study
movement preparation and execution. Frontiers in
neurology, 9:822.
Li, Y., Pan, Q., Wang, S., Peng, H., Yang, T., and Cambria,
E. (2019b). Disentangled variational auto-encoder
for semi-supervised learning. Information Sciences,
482:73–85.
Loshchilov, I. and Hutter, F. (2019). Decoupled weight
decay regularization. In International Conference on
Learning Representations.
Magnuson, J. R. and McNeil, C. J. (2021). Low-frequency
neural activity at rest is correlated with the movement-
related cortical potentials elicited during both real
and imagined movements. Neuroscience Letters,
742:135530.
Manjunatha, H. and Esfahani, E. T. (2021). Extracting in-
terpretable eeg features from a deep learning model to
assess the quality of human-robot co-manipulation. In
2021 10th International IEEE/EMBS Conference on
Neural Engineering (NER), pages 339–342. IEEE.
Olivas, B. E. and Chacon, M. (2018). Classification of mul-
tiple motor imagery using deep convolutional neural
networks and spatial filters. Applied Soft Computing,
75.
Pfurtscheller, G., Brunner, C., Schl
¨
ogl, A., and Da Silva,
F. L. (2006). Mu rhythm (de) synchronization and
EEG single-trial classification of different motor im-
agery tasks. NeuroImage, 31(1):153–159.
Riyad, M., Khalil, M., and Adib, A. (2021). MI-EEGNET:
A novel convolutional neural network for motor im-
agery classification. Journal of Neuroscience Meth-
ods, 353:109037.
Sakhavi, S., Guan, C., and Yan, S. (2018). Learning
temporal information for brain-computer interface us-
ing convolutional neural networks. IEEE Trans-
actions on Neural Networks and Learning Systems,
29(11):5619–5629.
Scapin, D., Cisotto, G., Gindullina, E., and Badia, L.
(2022). Shapley value as an aid to biomedical ma-
chine learning: a heart disease dataset analysis. In
2022 22nd IEEE International Symposium on Cluster,
Cloud and Internet Computing (CCGrid), pages 933–
939. IEEE.
Schirrmeister, R. T., Springenberg, J. T., Fiederer, L. D. J.,
Glasstetter, M., Eggensperger, K., Tangermann, M.,
Hutter, F., Burgard, W., and Ball, T. (2017). Deep
learning with convolutional neural networks for EEG
decoding and visualization. Human Brain Mapping.
Schneider, T., Wang, X., Hersche, M., Cavigelli, L., and
Benini, L. (2020). Q-EEGNet: An energy-efficient
8-bit quantized parallel EEGNet implementation for
edge motor-imagery brain-machine interfaces. In
2020 IEEE International Conference on Smart Com-
puting (SMARTCOMP), pages 284–289. IEEE.
vEEGNet: A New Deep Learning Model to Classify and Generate EEG
251

Zancanaro, A., Cisotto, G., Paulo, J. R., Pires, G., and
Nunes, U. J. (2021). CNN-based approaches for cross-
subject classification in motor imagery: From the
state-of-the-art to DynamicNet. In 2021 IEEE Confer-
ence on Computational Intelligence in Bioinformatics
and Computational Biology (CIBCB), pages 1–7.
Zancanaro, A., Cisotto, G., Tegegn, D. D., Manzoni, S. L.,
Reguzzoni, I., Lotti, E., and Zoppis, I. (2022). Vari-
ational autoencoder for early stress detection in smart
agriculture: A pilot study. In 2022 IEEE Workshop
on Metrology for Agriculture and Forestry (MetroA-
griFor), pages 126–130. IEEE.
Zoppis, I., Zanga, A., Manzoni, S., Cisotto, G., Morreale,
A., Stella, F., and Mauri, G. (2020). An attention-
based architecture for eeg classification. In BIOSIG-
NALS, pages 214–219.
ICT4AWE 2023 - 9th International Conference on Information and Communication Technologies for Ageing Well and e-Health
252
