
Speech Detection of Real-Time MRI Vocal Tract Data
Jasmin Menges
1
, Johannes Walter
2
, Jasmin Bächle
3
and Klemens Schnattinger
3
1
iRIX Software Engineering AG, Dornacherstrasse 192, 4053 Basel, Switzerland
2
Fraunhofer-Institut fuer Kurzzeitdynamik, Ernst-Mach-Institut, Am Klingelberg 1, 79588 Efringen-Kirchen, Germany
3
Business Innovation Center, Baden-Wuerttemberg Cooperative State University (DHBW),
Hangstrasse 46-50, Loerrach, Germany
schnattinger@dhbw-loerrach.de
Keywords: Deep Learning, Speech Production, MRI Data.
Abstract: This paper investigates the potential of Deep Learning in the area of speech production. The purpose is to
study whether algorithms are able to classify the spoken content based only on images of the oral region. With
the real-time MRI data of Lim et al. more detailed insights into the speech production of the vocal tract could
be obtained. In this project, the data was applied to recognize spoken letters from tongue movements using a
vector-based image detection approach. In addition, to generate more data, randomization was applied. The
pixel vectors of a video clip during which a certain letter was spoken could then be passed into a Deep
Learning model. For this purpose, the neural networks LSTM and 3D-CNN were used. It has been proven
that it is possible to classify letters with an accuracy of 93% using a 3D-CNN model.
1 INTRODUCTION
Developments in real-time MRI imaging have made
it possible to gain a more detailed insight into human
speech production. RT-MRI data of the vocal tract
can answer important questions in linguistics, speech
modelling, and clinical research. These data are the
basis for research regarding the ability to recognize
letters based on tongue movement. The research
question addressed in the paper is whether it is
possible to recognise the spoken content from an MRI
video of the oral cavity using Deep Learning. With
this information, a variety of in-depth research can be
initiated. For example, in linguistics to study different
movement patterns in different languages or in speech
modelling and clinical studies to compare patients
with speech problems using the deep learning model
to identify differences from typical movement
patterns and narrow down the problem to small sub-
areas of the oral cavity.
The dataset used includes MRI videos of vowels
and sentences from a total of 75 individuals with
different native languages. In this project the vowels
“a”, “e” and “o” of 5 different subjects were
considered. During the preparation of the data, the
timestamps for each spoken letter were analysed and
noted in an excel notebook. Furthermore, the pixel
data from the H5 files were merged with the excel
files and important parts of the videoframe were
extracted with the help of vectors. Training was first
performed with an LSTM model, a 3D CNN model
and later with a combination of both models. The
models were selected to evaluate the power of Deep
Learning for the application of complex data. The
obtained results were compared to identify the variant
with the highest accuracy (Lim et al. 2021).
2 DEEP LEARNING WITH
NEURAL NETWORKS
Artificial neural networks are versatile, powerful and
scalable, this makes them ideal for large and highly
complex machine learning tasks, including three-
dimensional data such as medical images and videos
(Géron 2019).
Two popular types for the use of deep learning in
the context of medical images are convolutional
neural networks (CNN) and recurrent neural
networks (RNN). In this study the focus is on 3D-
CNNs and LSTMs. CNNs have played a special role
in image recognition since 1980, and this has
increased considerably in recent years due to the
further development of computing power (Lu et al.
182
Menges, J., Walter, J., Bächle, J. and Schnattinger, K.
Speech Detection of Real-Time MRI Vocal Tract Data.
DOI: 10.5220/0012155600003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 1: KDIR, pages 182-187
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
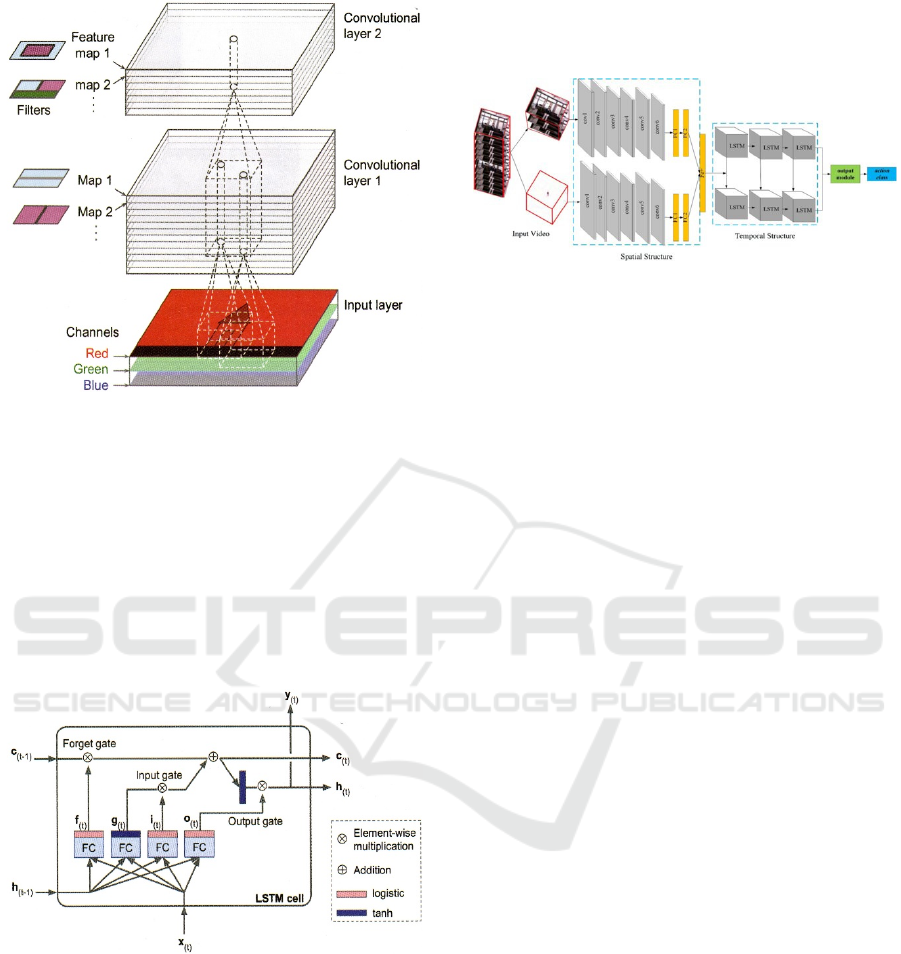
Figure 1: Convolutional layers with multiple feature maps,
and images with three colour channels (Géron 2019, p.452).
2019; Géron 2019). 3D-CNNs are a type of neural
network that have been adapted from the popular 2D-
CNNs for image analysis. In 3D-CNNs,
convolutional filters are applied to each 3D voxel of
the input volume, allowing for the detection of local
features and patterns within the volume. These
features can be combined and processed by
subsequent layers to extract more complex features
and classify the volume (cf. Figure 1).
Figure 2: LSTM cell (Géron 2019, p.516).
LSTM networks, on the other hand, are a type of
RNNs that are designed to process sequential data,
such as time series or text. LSTMs are capable of
learning long-term dependencies in the data and are
particularly effective in processing data with
temporal dynamics. In an LSTM network,
information is passed through a sequence of hidden
states, where each state has a memory cell and three
gating units: input gate, forget gate, and output gate
(cf. Figure 2). The gates control the flow of
information into and out of the memory cell, allowing
the network to selectively remember or forget
information from previous time steps (Ouyang et al.
2019).
Figure 3: Example of a basic architecture of a 3D-CNN-
LSTM (Jing et al. 2020).
In recent years, there has been growing interest in
combining 3D-CNNs and LSTM networks for
processing 3D sequential data, such as videos (cf.
Figure 3). This combination is known as 3D-CNN-
LSTM or spatiotemporal neural networks. In this
architecture, the 3D-CNN is used to extract spatial
features from each frame of the video, and the LSTM
network is used to process the temporal dynamics of
the video frames. The extracted features are then
combined and processed by subsequent layers to
classify the video. This approach has been shown to
be effective in various applications such as action
recognition, video captioning, and human pose
estimation (Jing et al. 2020).
3 FINDING AND
UNDERSTANDING DATA
The first step in realizing our Project was finding a
suitable data source. While MRI recordings are
relatively common, locating a dataset of freely
available MRI recordings was a challenge. Many
sources, such as the one from the Max Planck Society
on YouTube (MaxPlanckSociety 2018; Lim et al.
2021), only offer previews of single files.
Fortunately, Lim et al provided a large dataset
consisting of numerous MRI recordings of people
speaking. However, this presented our first hurdle:
understanding the dataset. The complete dataset is
available as a single zip archive, which is
approximately 550 gigabytes in size. It contains 3D
MRI scans, MP4 files containing multiple spoken
vowels, words or sentences, additional MP3 files with
the same audio data, as well as raw data of the MRI
scans, presented as 3-dimensional arrays in h5 files.
After obtaining the dataset, our next step was to
prepare it for use. Initially, we needed to understand
the data, including determining which files were
Speech Detection of Real-Time MRI Vocal Tract Data
183

Figure 4: Tree view of the dataset.
required and how to utilize them effectively. Figure 4
presents the tree structure of the dataset, where the
root folder consists of 75 subjects and three content
description files:
Subjects.xlsx
This file includes comprehensive demographic
information on the subjects, such as gender, age,
and spoken languages.
Stimuli.pptx
This file is a presentation that showcases all the
spoken letters, words and sentences present in
the dataset.
Metafile_public_202110129.json
This file not only provides demographic information
but also includes additional details about each
recording, such as audio quality.
Furthermore, as shown in Figure 4, each subject
has its folder, containing all scans and related data.
The t2w subfolders, which contain a massive dataset
of ".dicom" files, are nested within each subject's
folder. These files collectively generate a 3D scan for
each axis (x, y, and z), producing a clear view of the
head but providing no aid in recognizing spoken
letters.
The 3D subfolder includes two additional
subfolders, "recon" and "snapshot." The "snapshot"
subfolder contains numerous images, each
showcasing the subject in a particular position. On the
other hand, the files in the "recon" subfolder always
have a corresponding name, but they are in ".mat"
format, requiring MATLAB to access them. These
files contain raw pixel data in a 3D matrix, which
captures the subject's state at a single moment but
does not aid in the spoken letter recognition task.
Lastly, the 2drt folder contains audios of spoken
sequences, videos that combine the audio with a slice
of the MRI scan during speech, raw data from the
MRI scan in h5 format, and reconstructed data from
the raw data. The reconstructed data provides
information for the following machine learning
models, which aim to determine which letter is
spoken.
4 DATA PREPARATION
The "video" folder is especially useful in the data
preparation process, as each video contains multiple
spoken letters. To define when a letter is spoken, we
had to watch each video and record the exact time that
a subject began and ended speaking, as well as which
letter, they spoke. We created 13 different sequences
from five different speakers for this test. Table 1
depicts the head of one file.
One of the challenges we encountered in
identifying the exact time a subject began and ended
speaking was finding a media player capable of
handling such a high resolution. Since spoken letters
last less than a second, we required a player that could
display milliseconds and allow for frame-by-frame
stepping. We opted to use the MPC-HC-Player
Table 1: Preview of the csv data describing spoken letters in one video file.
Letter Letter Number Timestamp start Timestamp end
A
1 00:01.702 00:02.182
A 2 00:02.486 00:02:843
O 1 00:03.657 00:03.887
O 2 00:04.265 00:04.808
E 1 00:05.533 00:05.950
E 2 00:06.205 00:06.539
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
184

(MPC-HC Team 2020), which, although no longer
under development, could be easily configured to
meet our needs.
After obtaining the dataset, the next step was to
prepare it for use with machine learning models.
Leveraging our knowledge of Python, we used the
h5py library to read the files. Given that the MRI
scans were recorded at a frame rate of 83.28 frames
per second, the resulting files were usually over 100
MB in size. To reduce the amount of RAM required
to use the data, we read in each spoken letter
separately and discarded the sequences between
spoken parts.
In addition to reducing the amount of RAM
required to use the data, we also reduced the amount
of data transmitted to the machine learning models.
To achieve this, we followed the approach described
by Hellwig et al. in their paper and reduced the
dataset to a few vectors instead of using the entire
frame. To further increase the amount of training data
and reduce the risk of overfitting, we randomized the
placement of these vectors. This approach is similar
to using generated data to reduce bias in datasets.
Figure 4 demonstrates the process of pulling
pixels from the scan using vectors and how the
rotation of the vector results in subtle variations in the
training data. The left image in each set displays a
single frame of the scan extracted from the
reconstructed ("recon") data files, while the image on
the right shows the pixel values over time on the third
vector from the left. We utilized 7 vectors arranged in
a half circle to cover a substantial portion of the oral
cavity, as depicted in the images.
Table 2: Overall comparison of tested models.
Model La
y
ers Nodes Epochs Accurac
y
LSTM 3 33 10 66 %
LSTM 4 113 10 71 %
3D-CNN 5 123 5 93 %
3D-CNN + LSTM 6 143 5 90 %
Figure 5: Placement and rotation of the image vectors.
Speech Detection of Real-Time MRI Vocal Tract Data
185

5 DEEP LEARNING
IMPLEMENTATION
With the image-data for each spoken letter extracted,
we began implementing the machine learning layers.
We defined several smaller networks to test our
concept and compare different setups. Table 2
provides a brief overview of the models we used,
including the number of layers and nodes, the number
of epochs, and the accuracy achieved:
We tested different combinations of layers and
nodes and trained each model for a different number
of epochs. The best performing model was the 3D-
CNN, which achieved an accuracy of 93% with just 5
epochs. However, the 3D-CNN + LSTM model also
showed promising results, achieving an accuracy of
90% with 6 layers and 143 nodes.
Each model was implemented with Keras, an
open-source deep learning library. The first LSTM
model was a simple test that achieved great results in
tests with just a single subject. It consists of a network
with three layers, the first being a 20 LSTM nodes
layer, followed by a hidden layer of 10 densely
connected nodes. Finally, in every model, there are
three output nodes that return the calculated
probability of being each letter. Next, we tested a 3D-
CNN network consisting of five layers:
60 3D-convolutional nodes
30 3D-convolutional nodes
20 3D-convolutional nodes
10 dense nodes
3 output nodes
The increased training time and fast convergence
on the final accuracy led to the reduction to just five
epochs.
After this, we tried to combine the 3D-CNN with
the LSTM network by adding another layer between
layers 3 and 4 of the 3D-CNN model. This new layer
consists of 20 nodes and worsened the calculated
result compared to the simpler network. This could
have many reasons, such as a too small LSTM layer,
too little prepared training data, a bad integration of
the new nodes.
6 DISCUSSION AND LIMITS
In this paper it was studied whether it is possible to
recognize speech based on MRI videos with the help
of Deep Learning. Despite a positive result, there are
some points that need to be critically addressed.
1. Currently, only a limited number of people in
the dataset were examined, as data preparation
is manual and very time-consuming. The model
performs better with a lot of data from a few
people than with data from many different
people, because the variances naturally increase
with the number of different subjects. With the
option of adding multiple data from different
subjects to make the model more realistic, the
number of training data increases, which brings
us to the next point.
2. The 3D-CNN-LSTM model requires high
computing power. With the increase in training
data, the execution time continues to increase,
and powerful computers are needed.
3. Finally, it should be mentioned that only one
approach to detection was used in this paper,
which deals with the use of vectors. But there
are other approaches such as contour tracking.
This method defines outlines, or contours, of the
tongue and vocal tract which requires more
complex data preparation but could be more
precise through inclusion of all movements of
the oral cavity.
7 CONCLUSION
The investigations carried out have shown that it is
possible to recognize individual letters by using a
classification method. The best results were achieved
with the 3D-CNN model, which has an accuracy
value of 93%. The combination of the 3D-CNN
model and the LSTM model achieved a value of 90%.
The lower accuracy is because too little data available
for this solution.
8 OUTLOOK
For the future, the first step is to massively increase
the dataset to achieve better results. Therefore, it is
important to include more letters in the analysis.
Should this be successful, it would be conceivable to
analyse whole words. Above all, this project should
be an inspiration for anyone interested in further
research to achieve new insights in linguistics,
language modelling or clinical research.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
186

REFERENCES
Géron, A. (2019). Hands-on machine learning with Scikit-
Learn, Keras, and TensorFlow. Concepts, tools, and
techniques to build intelligent systems. Second edition.
Beijing, Boston, Farnham, Sebastopol, Tokyo: O'Reilly.
HDF5 for Python (2015). Available online at
https://www.h5py.org/, last updated 14.01.2015, last
checked 03.04.2023.
Jing, C., Wei, P., Sun, H. & Zheng, N. (2020).
Spatiotemporal neural networks for action recognition
based on joint loss. In: Neural Computing &
Applications 32 (9), 4293–4302. DOI: 10.1007/s00521-
019-04615-w.
Lim, Y., Toutios, A., Bliesener, Y., Tian, Y., Lingala, S. G.;
Vaz, C. & et al. (2021): A multispeaker dataset of raw
and reconstructed speech production real-time MRI
video and 3D volumetric images. In: Scientific data 8
(1), 187. DOI: 10.1038/s41597-021-00976-x.
Lu, H.; Wang, H.; Zhang, Q.; Yoon, S. W. & Won, D.
(2019): A 3D Convolutional Neural Network for
Volumetric Image Semantic Segmentation. In:
Procedia Manufacturing 39, 422–428. DOI:
10.1016/j.promfg.2020.01.386.
MaxPlanckSociety (2018): Echtzeit-MRT-Film: Sprechen -
YouTube. Available online at https://www.
youtube.com/watch?v=6dAEE7FYQfc, last updated
24.04.2018, last checked 02.04.2023.
MPC-HC Team (2020): MPC-HC Website. Available
online at https://mpc-hc.org/, last updated 15.09.2020,
last checked 03.04.2023.
Ouyang, X., Xu, S., Zhang, C., Zhou, P., Yang, Y., Liu, G.
& Li, X. (2019): A 3D-CNN and LSTM Based Multi-
Task Learning Architecture for Action Recognition. In:
IEEE Access 7, 40757–40770. DOI:
10.1109/ACCESS.2019.2906654. (HDF5 for Python
2015)
Speech Detection of Real-Time MRI Vocal Tract Data
187
