
BioSTransformers for Biomedical Ontologies Alignment
Safaa Menad
a
, Wissame Laddada
b
, Sa
¨
ıd Abdedda
¨
ım
c
and Lina F. Soualmia
d
Univ. Rouen Normandie, LITIS UR4108, 76000, Rouen, France
Keywords:
Language Models, Transformers, Siamese Neural Models, Zero-Shot Learning, Biomedical Texts, Biomedical
Ontologies, Ontology Alignment.
Abstract:
This paper aims at describing the new siamese neural models that we have developed. They optimize a self
supervised contrastive learning function on scientific biomedical literature articles. The results obtained on
several benchmarks show that the proposed models are able to improve various biomedical tasks without
examples (zero shot) and are comparable to biomedical transformers fine-tuned on supervised data specific to
the problems addressed. Moreover, these new siamese models are exploited to align biomedical ontologies,
demonstrating their semantic mapping capabilities. We then compare the different approaches of alignments
that we have proposed. In conclusion, we propose a distinct methods and data sources that we evaluate and
compare to validate our alignments.
1 INTRODUCTION
Ontology alignment plays a critical role in knowl-
edge integration. It aims at matching semantically re-
lated entities from different ontologies. Real-world
ontologies often contain a large number of classes,
which not only causes scalability issues, but also
makes it harder to distinguish classes with similar
names and/or contexts but representing different ob-
jects. Usual ontology alignment solutions typically
use lexical matching as their basis and combine it with
structural matching and logic-based mapping repair.
Recently, machine learning-based methods have
been proposed as alternative ways for lexical and
structural matching. For example, DeepAlignment
(Kolyvakis et al., 2018) relies on word embeddings
to represent classes and compute two classes’ sim-
ilarity according to their word vectors’ Euclidean
distance. Nevertheless, these methods adopt tradi-
tional non-contextual word embedding models such
as Word2Vec. Pre-trained transformer-based lan-
guage representation models such as BERT (Devlin
et al., 2019) can learn robust contextual text em-
beddings, and usually require only moderate train-
ing resources for fine-tuning. Although these mod-
els perform well in many Natural Language Process-
a
https://orcid.org/0009-0009-2204-7786
b
https://orcid.org/0000-0001-6841-7636
c
https://orcid.org/0000-0002-7521-7955
d
https://orcid.org/0000-0001-7668-2819
ing (NLP) tasks, they have not yet been sufficiently
investigated in ontology alignment tasks and concept
mapping.
The massive available biomedical data, such as
scientific articles, has also made it possible to train
these models on corpora for biomedical applications
(Alsentzer et al., 2019; Lee et al., 2020; Liu et al.,
2021). However, these language models require fine-
tuning on precise and rarely available supervised data
for each task, which strongly limits their use in prac-
tice. Since most biomedical NLP tasks (e.g., relation
extraction, document classification, question answer-
ing) can be reduced to the computation of a semantic
similarity measure between two texts (e.g., catego-
ry/article summary, query/results, question/answer),
we propose here to build new pre-trained siamese
models that embed pairs of semantically related texts
in the same vector representation space, and then
measure the similarity between them.
In this paper, we also bring transformers to the
ontology alignment task by (i) detailing our mod-
els BioSTransformers and BioS-MiniLM capable of
solving several NLP tasks without examples (zero
shot); (ii) showing experimentally on several biomed-
ical benchmarks that without fine-tuning for a specific
task, comparable results with biomedical transform-
ers fine-tuned on supervised data can be obtained;
and (iii) presenting how these models could be used
in order to semantically map entities from different
biomedical ontologies; and finally, evaluating our dif-
Menad, S., Laddada, W., Abdeddaïm, S. and Soualmia, L.
BioSTransformers for Biomedical Ontologies Alignment.
DOI: 10.5220/0012188600003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 2: KEOD, pages 73-84
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
73

ferent approaches of alignment and discussing the
validation of our results.
2 RELATED WORK
Several domain and application ontologies are used
for the same purpose. However, redundancy and
missing links between concepts from different on-
tologies may occur due to the heterogeneity of on-
tology modeling. In the literature, ontology align-
ment is proposed to overcome this heterogeneity and
allows semantic interoperability. In fact, consider-
ing an application, ontology alignment can be defined
as a semantic enhancement between concepts, roles,
and instances from several ontologies. In (Zimmer-
mann and Euzenat, 2006), the authors defined a dis-
tributed system as a system interconnecting two on-
tologies. Considering this definition, three seman-
tics of a distributed system are specified: simple dis-
tributed semantics where knowledge representation is
interpreted in one domain; integrated distributed se-
mantics where each local knowledge representation is
interpreted in its own domain; and contextualized dis-
tributed semantics where there is no global domain of
interpretation. In this paper, since we want to align
two ontologies from a single domain (biomedical on-
tologies) by means of pre-trained transformers, we
consider simple distributed semantics.
Ontology alignment results from an important
task known as the Ontology Matching (OM) where
a matcher is developed to identify similarities be-
tween ontologies. With regards to the classification
of matching systems presented in (Shvaiko and Eu-
zenat, 2013), a matcher can be based on terminologi-
cal (e.g., labels, comments, attributes, etc), structural
(ontology description), extensional (instances), or se-
mantics (interpretation and logic reasoning) similari-
ties. Moreover, because of the low level of semantic
expressiveness of some ontologies, external resources
can be exploited in the matching approaches.
It was for example the case in (Mary et al., 2017)
when they align the SNOMED CT with BioTopLite2
an upper level ontology.
Considering OM, an extensive survey is pre-
sented in (Portisch et al., 2022) to describe this ex-
ternal background knowledge and its usage. Fur-
thermore, the authors distinguish four categories of
matching approaches using background knowledge:
factual queries, where the data stored in the back-
ground knowledge is simply requested; structure-
based approaches, where structural elements in the
background knowledge are exploited; statistical/neu-
ral approaches (Fine-TOM (Hertling et al., 2021),
DAEOM (Wu et al., 2020)), where statistics or deep
learning are applied on the background knowledge;
and logic-based approaches where reasoning is em-
ployed with the external resource. For example,
(Chua and jae Kim, 2012) terminological, struc-
tural with background knowledge based on statistical
strategies were employed to map biomedical ontolo-
gies. Like CIDER-LM (Vela and Gracia, 2022), our
matching system relies on terminological similarities
with neural approaches to propagate a similarity con-
text between elements (properties and classes) from
two biomedical ontologies. The main difference be-
tween the two approaches is the embedding model
used. In (Vela and Gracia, 2022), they used the S-
BERT(Reimers and Gurevych, 2019) model, whereas
in our work we apply the BioSTransformers models
that we have developed.
3 TRANSFORMERS
Transformers are neural networks based on the multi-
head self-attention mechanism that significantly im-
proves the efficiency of training large models. They
consist of an encoder that transforms the input text
into a vector and a decoder that transforms this vector
into output text. The attention mechanism performs
better in these models by modeling the links between
the input and output elements. A pre-trained language
model (PLM) is a neural network trained on a large
amount of un-annotated data in an unsupervised way.
The model is then transferred to a target NLP task
(downstream task), where a smaller task-specific an-
notated dataset is used to fine-tune the PLM and to
build the final model capable of performing the target
task. The process is called fine-tuning a PLM.
3.1 Pre-Trained Language Models
Pre-trained language models such as BERT (Devlin
et al., 2019) have led to impressive gains in many
NLP tasks. Existing work generally focuses on gen-
eral domain data. In the biomedical domain, pre-
training on PubMed texts leads to better performance
in biomedical NLP tasks (Beltagy et al., 2019; Lee
et al., 2020; Peng et al., 2019). The standard approach
to pre-training a biomedical model starts with a gen-
eralized model and then follows by pre-training using
a biomedical corpus. For this purpose, BioBERT(Lee
et al., 2020) uses abstracts retrieved from PubMed and
full-text articles from PubMed Central (PMC). Blue-
BERT (Peng et al., 2019) uses both PubMed text and
MIMIC-III (Medical Information Mart for Intensive
Care) clinical notes (Johnson et al., 2016). SciBERT
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
74

(Beltagy et al., 2019) is an exception; the pre-training
is done from scratch, using the scientific literature.
3.2 Siamese Models
Sentence transformers have been developed to calcu-
late a similarity score between two sentences. They
are models that use transformers for tasks related
to sentence pairs: semantic similarity between sen-
tences, information retrieval, sentence reformulation,
etc. These transformers are based on two architec-
tures: cross-encoders that process the concatenation
of the pair, and bi-encoders siamese models that en-
code each pair element in a vector.
Sentence-BERT (Reimers and Gurevych, 2019) is
a BERT-based bi-encoder for generating semantically
meaningful sentence embeddings that are used in tex-
tual similarity comparisons. For each input, the model
produces a fixed-size vector (u and v). The objective
function is chosen so that the angle between the two
vectors u and v is smaller when the inputs are similar.
The objective function uses the cosine of the angle:
cos(u, v) =
u.v
|u||||v||
, if cos(u, v) = 1, the sentences are
similar and if cos(u, v) = 0, the sentences have no se-
mantic link.
Other sentence transformers models have been de-
veloped (Gao et al., 2021; Wang et al., 2021; Co-
han et al., 2020), among them, MiniLM-L6-v25
1
is a
bi-encoder based on a simplified version of MiniLM
(Wang et al., 2020). This fast and small model
has performed well on different tasks for 56 corpora
(Muennighoff et al., 2022).
4 PROPOSED MODELS:
BioSTransformers AND
BioS-MiniLM
Siamese transformers perform well in the general do-
main, but not in specialized ones (such as the biomed-
ical). Here we propose new siamese models pre-
trained on the PubMed corpus. Siamese transform-
ers were originally designed to transform (similarly
sized) sentences into vectors. In our approach, we
propose to transform MeSH (Medical Subject Head-
ings) terms, titles, and abstracts of PubMed arti-
cles in the same vector space by training a siamese
transformer model on these data. We want to en-
sure a match space between the short text and the
long text in this vector. Therefore, our models are
trained with pairs of inputs (title, MeSH term) and
1
https://huggingface.co/sentence-transformers/all-Min
iLM-L6-v2
(abstract, MeSH term). Based on these data, we have
built two models: the first one is our siamese trans-
former (BioSTransformers) based on a transformer
pre-trained on biomedical data, and the second one
is a siamese transformer already pre-trained on gen-
eralized data (BioS-MiniLM).
BioSTransformers. To build BioSTransformers, we
were inspired by the Sentence-BERT (Reimers and
Gurevych, 2019) model by replacing BERT with
other transformers. We used transformers that have
been trained on biomedical data (bio-transformers) to
create siamese transformers by adding a pooling layer
and changing the objective function. The pooling
layer computes the average vector of the transformer’s
output vectors (token embeddings). The two input
texts pass successively through the transformer pro-
ducing two vectors u and v at the output of the pool-
ing layer, which are then used by the objective func-
tion. To do so, we selected the best bio-transformers
BlueBERT (Peng et al., 2019), PubMed BERT (Gu
et al., 2022), BioELECTRA (Kanakarajan et al.,
2021) and Bio ClinicalBERT (Alsentzer et al., 2019).
These models were trained on PubMed except for
BlueBERT and Bio ClinicalBERT, which were also
trained on clinical notes. As a result, we constructed
the subsequent sentence-transformer models: S-
BlueBERT, S-PubMedBERT, S-BioELECTRA, and
S-BioClinicalBERT.
BioS-MiniLM. In this model, we used a siamese
transformer pre-trained on general data and then
trained it on our data. Several general sentence-
transformer models already pre-trained are available.
They differ in size, speed, and performance. In
those which obtained the best performances, we used
MiniLM-L6-v2 (see section 3.2) which has been pre-
trained on 32 general corpora (Reddit comments,
S2ORC, WikiAnswers, etc.).
Objective Function. In a sentence transformer, su-
pervised data are represented by triplets (sentence 1,
sentence 2, similarity score between the two sen-
tences). In our case, since we do not have any score
for abstracts nor titles and their corresponding MeSH
terms, we considered that:
• an abstract, a title, and the MeSH terms associated
with the same article (identified by a PMID) are
similar, and the score is equal to 1;
• an abstract (or a title) with MeSH terms not as-
sociated with the same article are not similar, and
the score is therefore equal to 0.
We use a self-supervised contrastive learning objec-
tive function based on the Multiple Negative Ranking
Loss (MNRL) function in the Sentence-Transformers
BioSTransformers for Biomedical Ontologies Alignment
75

package
2
. The MNRL only needs positive pairs as
input (the title (or abstract) and a MeSH term associ-
ated with the article in our case). For a positive pair
(title i or abstract i, MeSH i), MNRL considers that
each pair (title i or abstract i, MeSH j) with i 6= j in
the same batch is negative. Since an article can be
associated with several MeSH terms, we ensured in
the batch generation that an abstract (or title) associ-
ated with a MeSH term in PubMed is never taken as a
negative pair.
5 EXPERIMENTS AND RESULTS
5.1 Experiments
At first, to test the different transformers and the ob-
jective function to choose, we used only titles and re-
duced the number of MeSH terms. We selected 1,402
MeSH terms and 3.79 million pairs (title, MeSH) and
used 18,940 articles with their titles and MeSH terms
for validation.
In the second step, once we selected the trans-
former models and the objective function, we eval-
uated our BioSTransformers and BioS-MiniLM mod-
els on the (title, MeSH) and (abstract, MeSH) pairs
generated from all MeSH terms used in PubMed.
Since using all pairs from the 35 million articles in
PubMed is unnecessary (the model stabilizes), we se-
lected 6.75 million pairs for fine-tuning. And 18,557
articles were used for validation.
The two NLP tasks and the data used are described
below:
1. Document classification: the Hallmarks of Can-
cer (HoC) corpus consists of 1,852 abstracts of
PubMed publications manually annotated by ex-
perts according to a taxonomy composed of 37
classes. Each abstract in the corpus is assigned
zero to several classes (Hanahan and Weinberg,
2000);
2. Question answering (QA):
(a) PubMedQA: a corpus for Question answering
specific to biomedical research. It contains a
set of questions and an annotated field indicat-
ing whether the text contains the answer to the
research question (Jin et al., 2019);
(b) BioASQ: a corpus that contains several QA
tasks with expert annotated data, including
yes/no, list, and summary questions. We fo-
cused on the ”yes/no” question type (task 7b)
(Nentidis et al., 2019).
2
https://www.sbert.net/docs/package\ reference/losses.
html\#multiplenegativesrankingloss
We consider the two tasks (document classification
and QA) as a text similarity problem in order to re-
trieve the closest results for each query. We consider
the k closest results for each query, where k is the
number of results attributed to the query by the expert.
The similarity between the query and the results is
measured by the cosine similarity between the query
vector and the result vectors. In a classification task,
the query is the category, and the results are the doc-
uments classified in that category. In a QA task, the
query is the question, and the results are an answer.
5.2 Results
We evaluated our models according to the F1 score
used in the benchmarks HoC (Hanahan and Weinberg,
2000), PubmedQA (Jin et al., 2019), and BioASQ
(Nentidis et al., 2019) in (Gu et al., 2022). The re-
sults obtained by our zero shot models are given in
Table 1.
The results indicate that across the HoC bench-
mark, all our models perform similarly, achieving an
acceptable f1 score of 50%. However, for the other
two benchmarks, our S-PubMedBERT model outper-
forms the rest, yielding the best results.
Table 2 contains the results obtained on the same
tasks by models that are explicitly fine-tuned on these
tasks (Gu et al., 2022). These models are fine-tuned
for each benchmark with the supervised data available
in each case. These results show that the proposed
models can solve these tasks in a comparable way to
biomedical models fine-tuned on supervised data spe-
cific to the addressed problems that we did not use in
our zero shot approach.
For the HoC benchmark, the results obtained by
our best model are far below the results obtained by
PubMedBERT+fine-tuning (0.499 vs. 0.823). This
may be explained by the fact that the models in (Gu
et al., 2022) were fine-tuned specifically for each task,
including document classification, by modifying the
model architecture and adding specific layers for each
case.
On the other hand, for the PubMedQA bench-
mark, the results obtained by our model (best S-
PubMedBERT) are better than those obtained by
BioBERT+fine-tuning (0.729 vs. 0.602). Finally,
for the BioASQ benchmark, the results obtained by
our best model are acceptable compared to the re-
sults obtained by the fine-tuned models, even though
PubMedBERT+fine-tuning gives better results (0.751
vs. 0.876). All this done without re-adapting the ar-
chitecture of our models for each task and without
fine-tuning them on the specific data of the mentioned
benchmarks.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
76
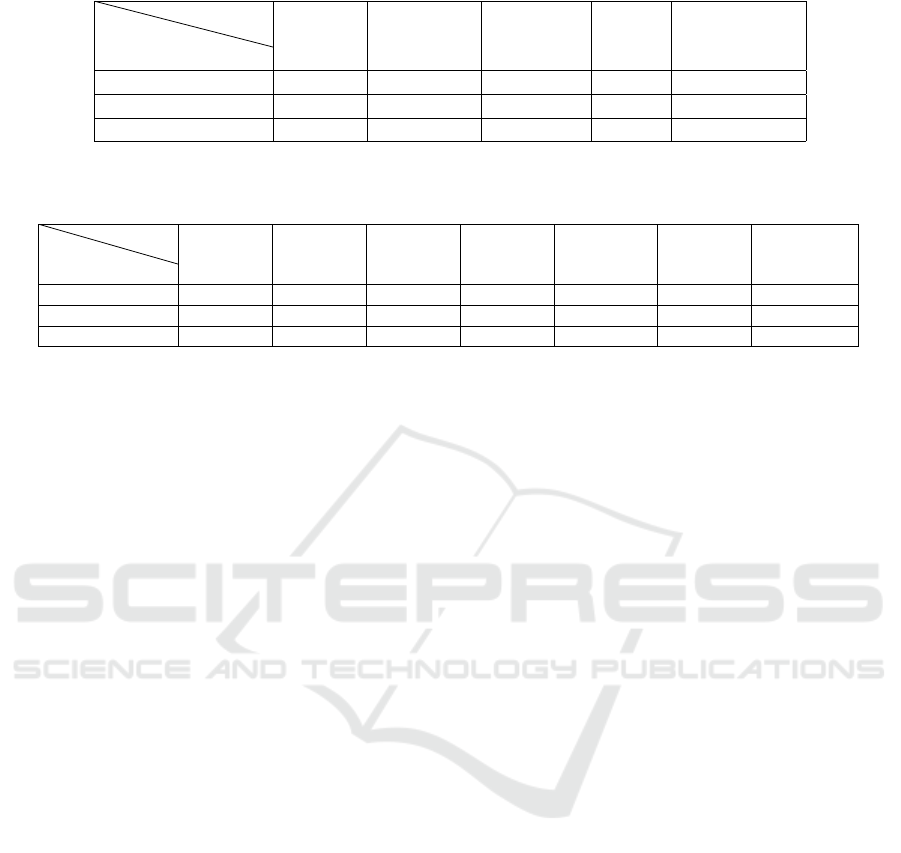
Table 1: Evaluation results (F1 score) of our models on different benchmarks.
Corpora
Model
BioS- S-Bio S-PubMed S-Blue S-BioClinical
MiniLM ELECTRA BERT BERT BERT
HoC 0.492 0.499 0.489 0.468 0.457
PubMedQA 0.649 0.675 0.729 0.652 0.652
BioASQ 0.747 0.694 0.751 0.713 0.714
Table 2: Evaluation results (F1 score) of the models fine-tuned specifically for these tasks on different benchmarks (Gu et al.,
2022).
Corpora
Model
BERT RoBERTa BioBERT SciBERT ClinicalBERT BlueBERT PubMedBERT
+fine-tuning +fine-tuning +fine-tuning +fine-tuning +fine-tuning +fine-tuning +fine-tuning
HoC 0.802 0.797 0.820 0.812 0.808 0.805 0.823
PubmedQA 0.516 0.528 0.602 0.574 0.491 0.484 0.558
BioASQ 0.744 0.752 0.841 0.789 0.685 0.687 0.876
Language models have gained widespread popu-
larity in NLP due to their ability to capture long-range
dependencies between words or concepts. This makes
them well-suited for tasks that require semantic un-
derstanding, such as ontology alignment. We have
leveraged the power of these models to improve align-
ment performance. Specifically, we apply our models
into an ontology alignment use case, in order to effec-
tively capture semantic similarities between concepts.
6 ONTOLOGY ALIGNMENT
TASK
This section is dedicated to the definitions inspired
from (Portisch et al., 2022; Euzenat et al., 2007; Os-
man et al., 2021). Although, we adapt these defini-
tions to our purpose, aligning two biomedical ontolo-
gies. Figure 1 summarizes the process of an ontology
matching following the definitions presented in this
section.
Ontology Definition: an ontology O
i
is a set of a vo-
cabulary defined by means of taxonomies to describe
a given domain of interest. This vocabulary is con-
sidered as a set of elements e
i
=< C
i
, R
i
, I
i
>; with
C
i
being the set of concepts, R
i
aggregates relations to
connect concepts, and I
i
gathers the set of instances to
interpret concepts and relate them with R
i
. An ontol-
ogy O
i
is also semantically enriched with X
i
to de-
fine axioms that formalize concepts based on logic
languages such as Description Logics or First Order
Logic.
Ontology Alignment and OM: an alignment de-
scribes the correspondence between two ontologies.
Formally, given two ontologies O
1
and O
2
, we limit
the definition of an alignment A to a set of triples.
Each triple is specified by the terminology of the bi-
nary relation r(e
1
, e
2
); where r depicts the relation
between the two elements e
1
∈ O
1
and e
2
∈ O
2
. Ac-
cordingly, the OM is the process of finding these sets
of correspondence. A confidence score c may also be
added to the correspondence triple to check the simi-
larity between e
1
and e
2
(e.g. the value of c ∈ [0,1]).
Matching System: it may be defined as a matching
function having several parameters to compute the
similarity between entities. F
m
(O
1
, O
2
, A
j
, P
c
, B) is a
matching function with P
c
as a parameter that holds
the confidence value of similarity and B the set of ex-
ternal resources used to find (or no) an alignment A
j
between the element e
1
and e
2
.
Ontology Integration: following the work presented
in (Osman et al., 2021), we define an ontology inte-
gration as a semantic enhancement of a target ontol-
ogy O
1
using elements from a source ontology O
2
.
The obtained result is a new ontology O
3
through the
alignment A =< r
j
, e
1, j
, e
2, j
, c
j
>.
7 ALIGNMENT MODELS
In this section, we describe our approach to align ele-
ments from different biomedical ontologies using our
previously described siamese models. Thus, the lat-
ter is a central system in the matching process. Since
transformers function as language models, it is neces-
sary that ontology elements are defined by labels (or
comments) and enriched by relations (properties).
We consider the matching process as a similar-
ity problem where our model (BioSTransformers) re-
ceives elements extracted from the input ontologies
and calculates their similarity. Based on the output
score, we conclude whether a match exists between
the two elements. Before delving into the details of
the approach deployed in our use case, we present the
BioSTransformers for Biomedical Ontologies Alignment
77

Figure 1: The matching process of ontologies (inspired from (Shvaiko and Euzenat, 2013)).
external ontologies used directly or indirectly in our
use case:
I) RxNorm (Nelson et al., 2011) is a standard nomen-
clature developed in the medical treatment field by the
NLM (United States National Library of Medicine).
The creation of this standard is motivated by the need
to unify the terminology used to represent drugs, as
well as to enable semantic interoperability. Addition-
ally, this standard provides normalization for clinical
drugs and related drug names. The latter are linked to
vocabularies commonly used in the same field.
II) ChEBI (Degtyarenko et al., 2008) is a dictio-
nary of molecular entities describing ”small” chem-
ical components (182,374 classes, 10 relations). The
molecular entities in question are either natural prod-
ucts or synthetic products. In addition to molecular
entities, ChEBI contains groups (part of molecular
entities) and entity classes. This dictionary thus in-
cludes an ontological classification, in which relations
between molecular entities or entity classes and their
parents and/or children are specified.
III) DRON (Hanna et al., 2013) was developed for in-
teroperability reasons and for the richness of seman-
tic expressiveness offered by ontologies. To achieve
this, the authors exploited external resources, namely,
RxNorm and ChEBI. Specifically, the development
of DRON is based on the alignment of entities from
RxNorm and entities from ChEBI. DRON is com-
posed of 661,999 classes and 125 relations with a
depth of 27 levels.
IV) DOID (Lynn et al., 2011) describes diseases and
medical vocabulary through the alignment of several
external resources. These vocabularies are used in, for
example, the annotation of biomedical data. Its cre-
ation is motivated by the need to represent knowledge
with semantic richness that allows linking biomedical
data on genes and diseases. It is composed of 8,127
classes, 46 relations, with a maximum depth of 13.
The use case describes the alignment of elements
from two biomedical ontologies: DOID (Human Dis-
ease Ontology
3
) and DRON (Drug Ontology
4
). The
result of this alignment represents an ontology inte-
gration in which each disease is associated with a list
of potential drugs.
To describe the approach of the alignment pro-
cess, the phases listed in (Osman et al., 2021) were
adopted.
7.1 Preprocessing Phase
Textual data was extracted from the two ontologies
DOID and DRON via SPARQL queries. This data is
related to: (i) the classes (element of DOID) that de-
scribe a disease
5
) and (ii) the metadata from ChEBI
(Chemical Entities of Biological Interest) from which
the DRON ontology was described. These metadata
represent information about a disease through a data
property definition (ChEBI metadata
6
). In BioPor-
tal, mappings have been established between (DOID)
and (DRON). However, these mappings only relate to
drugs that cause allergic reactions, rather than drugs
used to treat such reactions. Thus, there is currently
no association between DOID and DRON aimed at
proposing treatments for specific diseases. We were
able to extract a total of 13,678 diseases (DOID) and
3,295 metadata (DRON).
7.2 Matching Phase
The BioSTransformers model is used as the match-
ing function, where external knowledge bases repre-
sent the data on which the model is trained: first on
PubMed, and then on MIMIC III (a database contain-
ing electronic medical records of patients). For this
step, we chose the SBio ClinicalBERT model. Com-
pared to other models, this model provides good re-
3
https://bioportal.bioontology.org/ontologies/DOID
4
https://bioportal.bioontology.org/ontologies/DRON
5
http://purl.obolibrary.org/obo/
6
http://purl.obolibrary.org/obo/IAO
0
000115
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
78

sults for label comparison. This is due to the fact that
this model is trained on clinical notes from MIMIC
III.
7.3 Matching Process
To find similarities between disease names and meta-
data, we proceeded in different ways. First, we
took only the disease names from the DOID ontol-
ogy (rd f s : label) and calculated similarities between
these elements and the metadata of the DRON ontol-
ogy (obo : IAO
0
000115).
We then improved our process by considering two
approaches that take into account other elements of
DOID:
• The first one consists of concatenating several el-
ements of the DOID ontology. These elements
correspond to the name of the disease (rd f :
label), its definition (obo : IAO
0
000115), and
several synonymous disease names (oboInOwl :
hasExactSynonym). We call this strategy ”multi-
label”. The concatenation is considered as an in-
put for BioSTransformers.
• The second approach consists of considering only
one element at a time from DOID. Specifically,
we take into account either the name of the dis-
ease (rd f : label), or the definition of the disease
(obo : IAO
0
000115), or a single related disease
name (oboInOwl : hasExactSynonym) in each
similarity calculation. Thus, for each element
from DRON considered by BioSTransformers,
the correspondence is established with an element
from DOID, by choosing the maximum similar-
ity score between the metadata from DRON (obo :
IAO
0
000115) and one of the metadata from DOID
(rd f : label or oboInOwl : hasExactSynonym or
obo : IAO
0
000115). This score must be greater
than 0.5. We call it ”max-label”.
Figure 3 and Figure 4 describe the metadata extracted
from DOID and DRON.
7.4 Merging Phase
The generated alignments are correspondences be-
tween a single concept from DOID and a single con-
cept from DRON (one-to-one alignment). The type
of correspondence is an inclusion between the meta-
data that define a ChEBI class and those that define
a disease. This alignment is maintained when the
confidence score (similarity score) is higher than the
threshold of 0.5. We initially selected the threshold
value of 0.5 due to its intrinsic significance as the
midpoint, We plan to explore and assess performance
with threshold values below 0.5 to consider predic-
tions that may be slightly lower but still meaningful.
If an alignment exists, then a new relation is
defined between the disease and the ChEBI con-
cept. This new relation allows the generation of a
third ontology (integration ontology) enriched by the
DRON and DOID ontologies. We name this relation
Has Medicine with CHEBI. Figure 2 illustrates how
BioSTransformers are used in the ontology alignment
task.
The number of alignments generated by the three
approaches is reported in Table 3. One can observe
that the third approach produces the largest number
of alignments. Thus, the name of the disease is not as
representative as the other metadata.
The results obtained are very encouraging when
using BioSTransformers to find similarity. For exam-
ple, in DRON, the element ”CHEBI 31286”, which
composes the drug under the name ”bifonazole”,
is defined by the metadata ”A racemate compris-
ing equimolar amounts of R- and S-bifonazole. It
is a broad spectrum antifungal drug used for the
treatment of fungal skin and nail infections.”. In
DOID, the disease ”DOID
13074” is defined by the
metadata ”tinea unguium”. The matching process
gives a similarity score of 0.561. Since the con-
fidence score is greater than 0.5, we create a new
relation ”Has Medicine with CHEBI(DOID 13074,
CHEBI 31286)”. All new relations can be retrieved
through a simple SPARQL query.
7.5 Evaluations of the Alignments
The next necessary step is to evaluate and validate the
obtained alignments. For this purpose, we propose to
rely on the use of several knowledge bases, namely:
7.5.1 The UMLS Metathesaurus
(Unified Medical Language System)
7
as an external
evaluation resource. For each disease, we searched
for its corresponding drug in the UMLS using its CUI
(Concept Unique Identifier) and the UMLS API avail-
able at the following URL: https://uts-ws.nlm.nih.gov
/rest/content/current/CUI/code/relations?includeAd
ditionalRelationLabels=may be treated by&apiKey.
In this URL, code represents the CUI, and by utilizing
the semantic relation may be treated by, we retrieved
the treatment information. Table 4 shows the num-
ber of diseases that were associated with CUI codes
in our alignments. For the remaining diseases, alter-
native codes were required, which were not utilized
during the data extraction process.
7
https://www.nlm.nih.gov/research/umls/index.html
BioSTransformers for Biomedical Ontologies Alignment
79
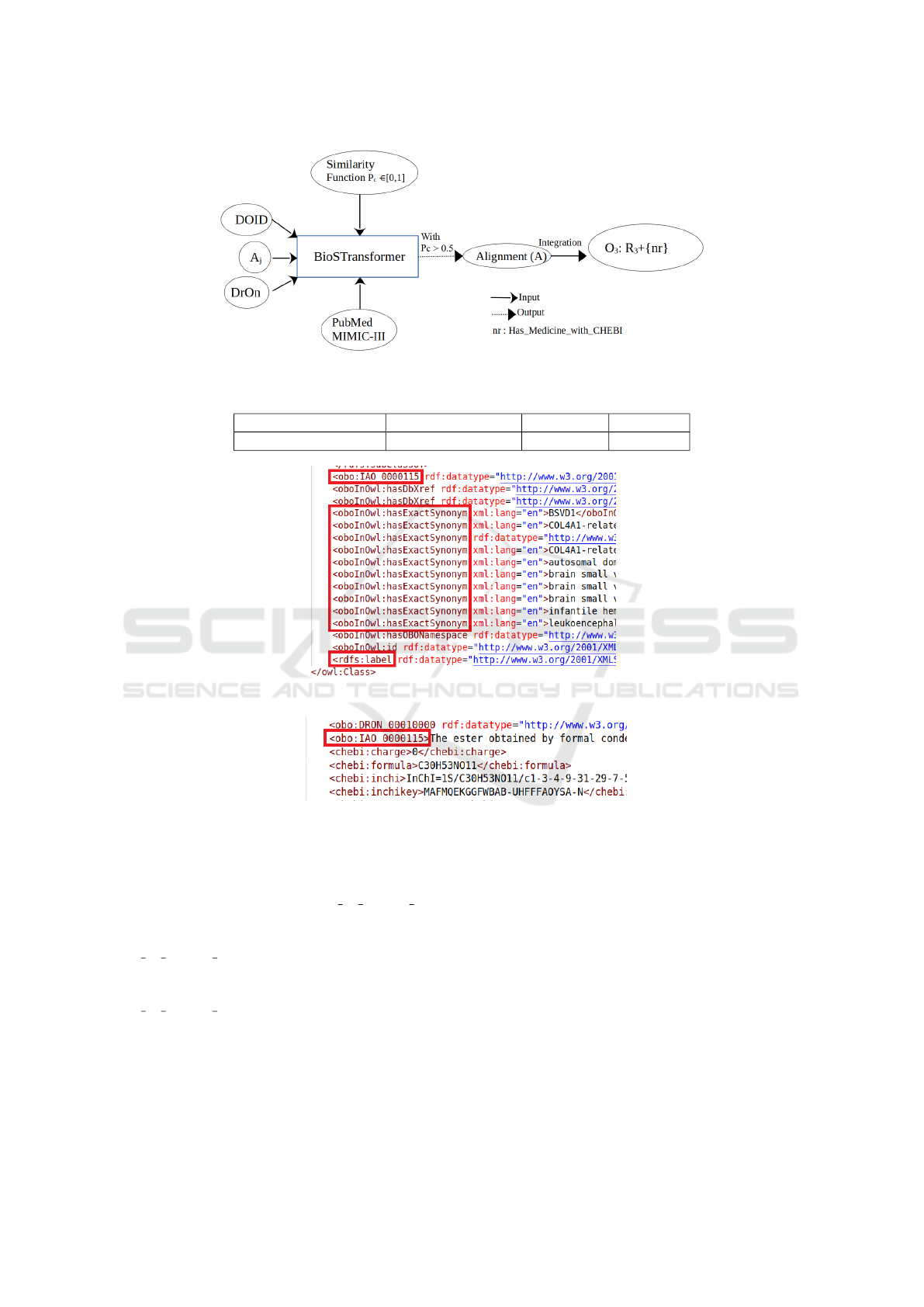
Figure 2: The matching process of DOID and DrOn using BioSTransformers.
Table 3: Number of alignments generated for each matching approach.
Approach Disease name only multi-label max-label
Alignment’s’ number 615 770 1,035
Figure 3: The relations considered by our model from DOID to calculate similarity.
Figure 4: The metadata considered by our model from DRON to calculate similarity.
For the diseases with CUI codes, we conducted
a search in UMLS to find their corresponding drugs.
However, we discovered that not all of them were re-
lated with the semantic relation may be treated by in
the UMLS Semantic Network. Table 4 displays the
number of diseases with CUI codes that also have the
may be treated by relation.
Therefore, we were only able to evaluate
the diseases that had both the CUI and the
may be treated by relation. During our evaluation,
we discovered several diseases in UMLS that had the
exact same drug as suggested by our models.
After analyzing the results more thoroughly and
carefully examining the definitions of each drug, we
concluded that the mismatches we encountered were
primarily a result of our model suggesting chemical
entities or agents that were components of the same
drug in UMLS. This discrepancy can be attributed
to the fact that we conducted the alignment using
DRON, which is based on ChEBI—an ontology of
chemical entities.
In response to the challenges faced, we endeav-
ored to explore alternative and robust methods for
validating the alignments with greater accuracy and
comprehensiveness. To achieve this objective, we in-
tegrated the OpenFDA into our work. This helped us
improve the precision and completeness of our align-
ment validations, allowing us to analyze the data more
effectively.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
80

Table 4: Evaluation’s results.
Diseases names umls
Approach
Disease name only multi-label max-label
with CUI 262 492 641
with CUI and the may be treated by relation 109 132 171
7.5.2 OpenFDA
OpenFDA
8
(Kass-Hout et al., 2016) is an initiative by
the U.S. Food and Drug Administration (FDA) that
provides public access to datasets and APIs related
to FDA-regulated products. It aims to promote data
transparency, facilitate research and analysis, moni-
tor product safety, and encourage application devel-
opment. OpenFDA serves as a valuable resource for
researchers, developers, healthcare professionals, and
the general public interested in FDA-generated data.
It offers many APIs: drug adverse events, med-
ical device adverse events, drug labels, device clas-
sifications, product recalls, and enforcement reports.
By querying these APIs, researchers can access in-
formation about drug adverse events, medical device
recalls, and food safety-related enforcement actions,
among other datasets. This rich and diverse collection
of data empowers researchers to conduct comprehen-
sive analyses and gain valuable insights into public
health trends and safety issues.
To address our research questions, we utilized the
drug labels API from https://open.fda.gov/apis/dr
ug/label/. Specifically, we used the following query:
https://api.fda.gov/drug/label.json?search=desc
ription:”Drug label”&limit=10. In this context,
the term description pertains to the targeted field we
aimed to retrieve, and Drug
label denotes the specific
drug name we search for.
Our search strategy involved looking for particu-
lar drug names by choosing the desired fields to ex-
amine, such as description, indications and usage etc.
This approach enabled us to study different informa-
tion and discover the most relevant for our study. The
results of this analysis helped us better understand the
drug-related aspects we were investigating.
Example:
Query:
htt ps :// api . fda . go v / drug / lab el .
json ? se arch = d esc ri ption :"
ox yt et ra cy cline "& lim it =10
Result:
" d es cri pt io n ": [
8
https://open.fda.gov/
" D ES CRI PT IO N Do xy cyc li ne
is an an ti ba cte ri al
drug s yn th et ica ll y
de riv ed fro m
ox yte tra cyc line , and
is avail abl e as
do xy cyc li ne h ycl ate
tabl ets , USP . The
st ru ctu ra l fo rmu la of
do xy cyc li ne
mo no hyd ra te is w ith a
mo lecul ar f orm ula ... "
],
" i nd ic at io ns _a nd _u sa ge ": [
" I ND ICA TI ON S AN D US AGE To
re duce the d ev el opm en t
of drug - res is tan t
ba cte ria and m ai nta in
ef fe ct ivene ss of
do xy cyc li ne and o ther
an ti ba cteri al drugs ,
do xy cyc li ne s hou ld be
used on ly to tre at or
pr eve nt i nfect ions
that are prove n or
st ron gly su spe cted to
be c aus ed by
su sc ept ib le b ac ter ia
... "
],
Upon retrieving the drug data, we proceeded to
conduct string matching (defined below) between dis-
ease names and the text present in these fields. This
approach aimed to identify corresponding disease
names within the descriptions, enabling us to assess
whether the drug is suitable for treating the specific
disease or not. By performing this comparison, we
sought to determine the potential efficacy of the drug
in addressing the targeted medical conditions, thereby
enhancing our understanding of its applicability in the
context of the disease.
String Matching. String matching, also known as
string searching, is a fundamental operation in com-
puter science and refers to the process of finding oc-
currences of a given pattern (a sequence of charac-
ters) within a longer text (a string). The goal of string
matching is to determine if the pattern exists in the
BioSTransformers for Biomedical Ontologies Alignment
81
text and, if so, identify the positions or indices where
the pattern occurs.
Despite using string matching or word-to-word
comparison to analyze the data, we found that this ap-
proach did not yield comprehensive results. The lim-
itations of this method became evident as it could not
provide a comprehensive understanding of the rela-
tionships between the disease names and the drug de-
scriptions. During our analysis, we occasionally en-
countered additional synonyms or extended names of
the disease. These variations in disease names added
complexity to the matching process and required fur-
ther consideration to ensure accurate and comprehen-
sive results.
To address this issue and obtain more accurate and
insightful outcomes, we explored alternative method-
ologies that could offer a more comprehensive and nu-
anced analysis of the data.
WordNet. As a result, we adapted our approach
to improve the analysis’s effectiveness in capturing
all pertinent disease information. We explored the
use of WordNet, a resource that offers disease syn-
onyms, to further enrich our analysis and ensure com-
prehensive coverage of disease-related terms. Word-
Net (Miller et al., 1990) is a lexical database and
semantic network for the English language. It was
created at Princeton University and is widely used
in various natural language processing (NLP) appli-
cations. WordNet organizes words into sets of syn-
onyms, called synsets, each representing a distinct
concept. These synsets are linked together through
semantic relationships such as hypernyms (more gen-
eral terms) and hyponyms (more specific terms).
The main purpose of WordNet is to provide a com-
prehensive and structured resource for understanding
the meanings of words and their relationships. It has
been used in various NLP tasks, such as word sense
disambiguation, text summarization, machine transla-
tion, and information retrieval.
Example:
Disease name: Lymphopenia
Wordnet synonyms: lymphocytopenia,
blood disorder, etc.
However, there were instances when even this
method proved insufficient. For instance, in the ex-
ample mentioned earlier, we could not find the dis-
ease name or its synonyms because the description
field contained the term ”lymphocytes,” which was
not explicitly mentioned in the disease name or in its
synonyms. To address this challenge, we explored al-
ternative methods to enhance the accuracy and com-
pleteness of our analysis in such cases.
Knowledge Representation Systems. To enhance
our analysis, we used the UMLS metathesaurus (http
s://www.nlm.nih.gov/research/umls/index.html),
which provides structured knowledge and relation-
ships between medical concepts. This resource al-
lowed us to compare medical terms based on their
hierarchical relationships, semantic similarity, and
shared attributes.
In the UMLS, each concept is categorized into one
or more Semantic Types, which are broad classifi-
cations representing different facets of the concept’s
meaning. The ”mother class” serves as a top-level
Semantic Type, encapsulating the most general cate-
gory to which the concept belongs. This hierarchi-
cal organization of Semantic Types helps in system-
atically grouping and understanding the various con-
cepts within the UMLS, making it easier to navigate
and extract relevant information from this extensive
lexical resource.
Therefore, we attempted to retrieve the mother
class of the disease concept to address instances
where disease names might not exactly match or lack
clarity. By using the broader and more clearly defined
mother class for comparisons, we facilitated the pro-
cess of matching and analyzing medical terms. This
approach made the comparisons much simpler and
more practical.
Example:
Disease name: Amyotrophic Lateral Sclero-
sis, Guam Form
Mother class: parent disorders of peripheral
nerve, neuromuscular junction and muscle
Table 5 presents the results obtained using the
proposed methods: string matching, WordNet, and
UMLS mother concepts, for each alignment ap-
proach: disease name only, multi-label, and max la-
bel. The scores are calculated as follows: -1 indicates
that the drug name does not exist in OpenFDA, possi-
bly due to discontinuation or replacement in the mar-
ket; 0 signifies that the disease name we are search-
ing for does not exist in the drug’s description in
OpenFDA; and 1 indicates that the disease name,
its synonym, or its mother concept is present in the
drug’s description in OpenFDA.
In summary, our approach involved the initial ap-
plication of the string matching method. For align-
ments with a score of -1, indicating a lack of data in
the current resource, we sought alternative sources or
sought expert assistance to validate the alignments.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
82
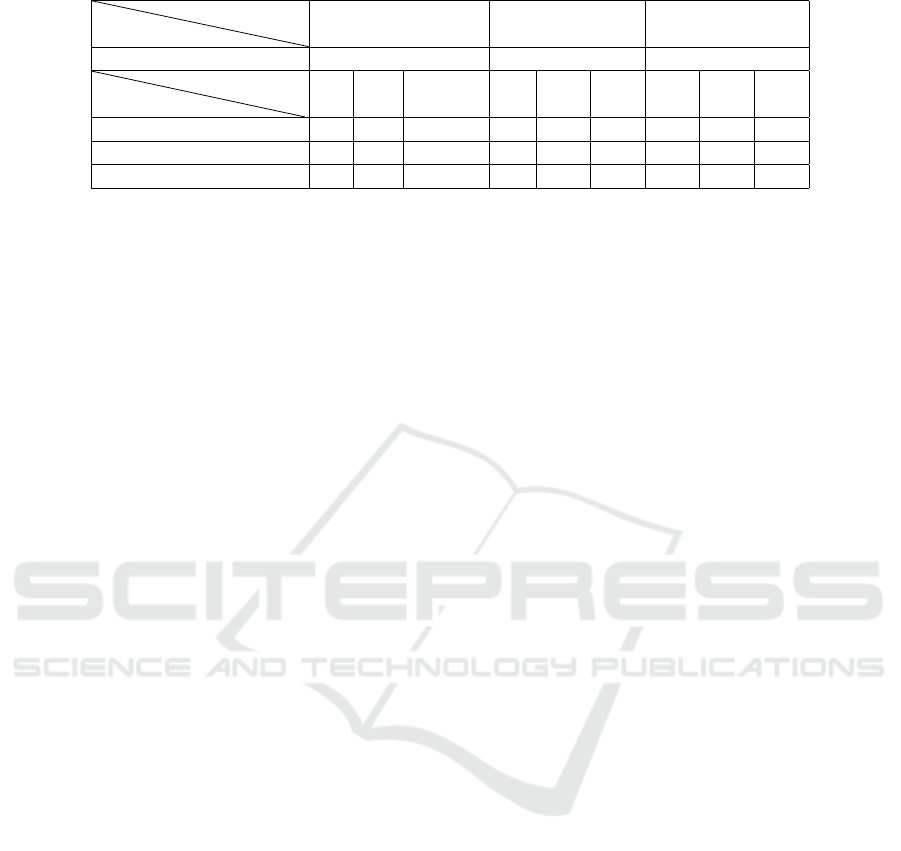
Table 5: Comparison of alignment results using different methods: String matching, WordNet, and UMLS mother concepts.
Disease
Approach
Disease name only multi-label max-label
with CUI 262 492 641
Method
Score me
-1 0 1 -1 0 1 -1 0 1
String matching 49 33 178 89 148 255 128 214 299
Wordnet 0 24 9 0 125 23 0 168 46
UMLS mother concept 0 13 20 0 73 75 0 100 114
For alignments with a score of 0, we selected them
and performed further validation using both WordNet
and UMLS mother concept methods. By employing
these additional techniques, we aimed to determine
the validity of these alignments. We plan to combine
these approaches and find others to make our analysis
more complete and accurate. By combining different
techniques, we aim to get better and more reliable re-
sults.
Alignments with a score of 1 are considered valid,
as they indicated that the disease name, its synonym,
or its mother concept was present in the drug’s de-
scription.
8 CONCLUSION
In this paper, we proposed new siamese models that
can improve the results of two biomedical NLP tasks
in a zero-shot context. These models embed pairs
of texts in the same representation space and calcu-
late the semantic similarity between texts of differ-
ent lengths. We then evaluated our models on several
biomedical benchmarks and showed that without fine-
tuning on a specific task, we achieved results compa-
rable to those of biomedical transformers fine-tuned
on task-specific supervised data. In addition, we pro-
posed to exploit our models in a practical scenario that
consists of aligning entities from two distinct biomed-
ical ontologies to establish new relations.
The evaluation of our alignments, based on these
results, has shown promising outcomes. Currently,
we are in the process of integrating and combining
additional data sources and methods to further vali-
date the remaining alignments. This ongoing valida-
tion process will enhance the reliability of our find-
ings, contributing to a more robust and accurate drug-
disease recommendation process. The integration of
other ontologies (e.g., adverse drug effects or other
drug resources like DrugBank) is planned as well as
the validation of the remaining alignments not found
in the available resources by experts. Furthermore,
we intend to assess the efficacy of our approach on
alignments involving domain ontologies.
This paper presents the initial outcomes of our re-
search project focused on the development of a diag-
nostic system that aims to create a diagnostic predic-
tion tool to enhance patient care. These alignments
will enable us to achieve semantic interoperability be-
tween health systems.
REFERENCES
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jindi,
D., Naumann, T., and McDermott, M. (2019). Pub-
licly available clinical BERT embeddings. In Pro-
ceedings of the 2nd Clinical Natural Language Pro-
cessing Workshop, pages 72–78, Minneapolis, Min-
nesota, USA. Association for Computational Linguis-
tics.
Beltagy, I., Lo, K., and Cohan, A. (2019). SciBERT: A
pretrained language model for scientific text. In Pro-
ceedings of the 2019 Conference on Empirical Meth-
ods in Natural Language Processing and the 9th Inter-
national Joint Conference on Natural Language Pro-
cessing (EMNLP-IJCNLP), pages 3615–3620.
Chua, W. W. K. and jae Kim, J. (2012). Boat: Auto-
matic alignment of biomedical ontologies using term
informativeness and candidate selection. Journal of
Biomedical Informatics, 45(2):337–349.
Cohan, A., Feldman, S., Beltagy, I., Downey, D., and Weld,
D. S. (2020). Specter: Document-level representation
learning using citation-informed transformers. In Pro-
ceedings of the 58th Annual Meeting of the Associa-
tion for Computational Linguistics, pages 2270–2282.
Degtyarenko, K., Matos, P., Ennis, M., Hastings, J.,
Zbinden, M., McNaught, A., Alc
´
antara, R., Darsow,
M., Guedj, M., and Ashburner, M. (2008). Chebi: A
database and ontology for chemical entities of biolog-
ical interest. Nucleic acids research, 36:D344–50.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). BERT: Pre-training of deep bidirectional
transformers for language understanding. In Proceed-
ings of NAACL-HLT, pages 4171–4186.
Euzenat, J., Shvaiko, P., et al. (2007). Ontology matching,
volume 18. Springer.
Gao, T., Yao, X., and Chen, D. (2021). Simcse: Simple con-
trastive learning of sentence embeddings. In Proceed-
ings of the 2021 Conference on Empirical Methods in
Natural Language Processing, pages 6894–6910.
BioSTransformers for Biomedical Ontologies Alignment
83

Gu, Y., Tinn, R., Cheng, H., Lucas, M., Usuyama,
N., Liu, X., Naumann, T., Gao, J., and Poon, H.
(2022). Domain-specific language model pretrain-
ing for biomedical natural language processing. ACM
Transactions on Computing for Healthcare, 3(1):1–
23.
Hanahan, D. and Weinberg, R. A. (2000). The hallmarks of
cancer. Cell, 100(1):57–70.
Hanna, J., Joseph, E., Brochhausen, M., and Hogan, W.
(2013). Building a drug ontology based on rxnorm
and other sources. Journal of biomedical semantics,
4:44.
Hertling, S., Portisch, J., and Paulheim, H. (2021). Match-
ing with transformers in melt.
Jin, Q., Dhingra, B., Liu, Z., Cohen, W., and Lu, X. (2019).
PubMedQA: A dataset for biomedical research ques-
tion answering. In Proceedings of (EMNLP-IJCNLP),
pages 2567–2577.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). MIMIC-
III, a freely accessible critical care database. Scientific
data, 3(1):1–9.
Kanakarajan, K. r., Kundumani, B., and Sankarasubbu, M.
(2021). BioELECTRA: Pretrained biomedical text en-
coder using discriminators. In Proceedings of the 20th
Workshop on Biomedical Language Processing, pages
143–154, Online. Association for Computational Lin-
guistics.
Kass-Hout, T. A., Xu, Z., Mohebbi, M., Nelsen, H., Baker,
A., Levine, J., Johanson, E., and Bright, R. A. (2016).
Openfda: an innovative platform providing access
to a wealth of fda’s publicly available data. Jour-
nal of the American Medical Informatics Association,
23(3):596–600.
Kolyvakis, P., Kalousis, A., and Kiritsis, D. (2018).
Deepalignment: Unsupervised ontology matching
with refined word vectors. In Proceedings of NAACL-
HLT, 787–798., pages 787–798.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). BioBERT: a pre-trained biomed-
ical language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Liu, F., Shareghi, E., Meng, Z., Basaldella, M., and Col-
lier, N. (2021). Self-alignment pretraining for biomed-
ical entity representations. In Proceedings of NAACL-
HLT, pages 4228–4238.
Lynn, S., Arze, C., Nadendla, S., Chang, Y.-W. W.,
Mazaitis, M., Felix, V., Feng, G., and Kibbe, W.
(2011). Disease ontology: A backbone for disease se-
mantic integration. Nucleic acids research, 40:D940–
6.
Mary, M., Soualmia, L., Gansel, X., Darmoni, S., Karls-
son, D., and Schulz, S. (2017). Ontological represen-
tation of laboratory test observables: Challenges and
perspectives in the snomed ct observable entity model
adoption. pages 14–23.
Miller, G. A., Beckwith, R., Fellbaum, C., Gross, D., and
Miller, K. J. (1990). Introduction to wordnet: An on-
line lexical database. International journal of lexicog-
raphy, 3(4):235–244.
Muennighoff, N., Tazi, N., Magne, L., and Reimers, N.
(2022). Mteb: Massive text embedding benchmark.
arXiv preprint arXiv:2210.07316.
Nelson, S. J., Zeng, K., Kilbourne, J., Powell, T., and
Moore, R. (2011). Normalized names for clinical
drugs: RxNorm at 6 years. Journal of the American
Medical Informatics Association, 18(4):441–448.
Nentidis, A., Bougiatiotis, K., Krithara, A., and Paliouras,
G. (2019). Results of the seventh edition of the
BioASQ challenge. In Joint European Conference
on Machine Learning and Knowledge Discovery in
Databases, pages 553–568. Springer.
Osman, I., Ben Yahia, S., and Diallo, G. (2021). Ontol-
ogy integration: Approaches and challenging issues.
Information Fusion, 71:38–63.
Peng, Y., Yan, S., and Lu, Z. (2019). Transfer learn-
ing in biomedical natural language processing: An
evaluation of BERT and ELMo on ten benchmarking
datasets. In Proceedings of the 18th BioNLP Work-
shop and Shared Task, pages 58–65, Florence, Italy.
Association for Computational Linguistics.
Portisch, J., Hladik, M., and Paulheim, H. (2022). Back-
ground knowledge in ontology matching: A survey.
Semantic Web, pages 1–55.
Reimers, N. and Gurevych, I. (2019). Sentence-BERT: Sen-
tence embeddings using Siamese BERT-networks. In
Proceedings of (EMNLP-IJCNLP), pages 3982–3992,
Hong Kong, China. Association for Computational
Linguistics.
Shvaiko, P. and Euzenat, J. (2013). Ontology matching:
State of the art and future challenges. IEEE Transac-
tions on Knowledge and Data Engineering, 25:158–
176.
Vela, J. and Gracia, J. (2022). Cross-lingual ontology
matching with cider-lm: results for oaei 2022.
Wang, K., Reimers, N., and Gurevych, I. (2021). Tsdae:
Using transformer-based sequential denoising auto-
encoderfor unsupervised sentence embedding learn-
ing. In Findings of the Association for Computational
Linguistics: EMNLP 2021, pages 671–688.
Wang, W., Wei, F., Dong, L., Bao, H., Yang, N., and Zhou,
M. (2020). Minilm: Deep self-attention distillation for
task-agnostic compression of pre-trained transform-
ers. Advances in Neural Information Processing Sys-
tems, 33:5776–5788.
Wu, J., Lv, J., Guo, H., and Ma, S. (2020). Daeom: A
deep attentional embedding approach for biomedical
ontology matching. Applied Sciences, 10(21).
Zimmermann, A. and Euzenat, J. (2006). Three semantics
for distributed systems and their relations with align-
ment composition. In The Semantic Web - ISWC 2006,
pages 16–29, Berlin, Heidelberg. Springer Berlin Hei-
delberg.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
84
