
Pleural Effusion Classification on Chest X-Ray Images with Contrastive
Learning
Felipe Andr
´
e Zeiser
1 a
, Ismael Garcia Santos
1
, Henrique Christoph Bohn
1
,
Cristiano Andr
´
e da Costa
1 b
, Gabriel de Oliveira Ramos
1 c
, Rodrigo da Rosa Righi
1 d
,
Andreas Maier
2 e
, Jos
´
e Rodrigo Mendes Andrade
3
and Alexandre Bacelar
3
1
Software Innovation Laboratory - SOFTWARELAB,
Universidade do Vale do Rio dos Sinos - Unisinos, S
˜
ao Leopoldo, Brazil
2
Department of Computer Science, Friedrich-Alexander-Universit
¨
at, Erlangen-N
¨
urnberg, Erlangen, Germany
3
Medical Physics and Radioprotection Service, Hospital de Clinicas de Porto Alegre, Porto Alegre, Brazil
Keywords:
Contrastive Learning, X-Ray, Pleural Effusion.
Abstract:
Diagnosing pleural effusion is important to recognize the disease’s etiology and reduce the length of hos-
pital stay for patients after fluid content analysis. In this context, machine learning techniques have been
increasingly used to help physicians identify radiological findings. In this work, we propose using contrastive
learning to classify chest X-rays with and without pleural effusion. A model based on contrastive learning is
trained to extract discriminative features from the images and reports to maximize the similarity between the
correct image and text pairs. Preliminary results show that the proposed approach is promising, achieving an
AUC of 0.900, an accuracy of 86.28%, and a sensitivity of 88.54% for classifying pleural effusion on chest
X-rays. These results demonstrate that the proposed method achieves comparable or superior to state of the art
results. Using contrastive learning can be a promising alternative to improve the accuracy of medical image
classification models, contributing to a more accurate and effective diagnosis.
1 INTRODUCTION
Early pleural effusion detection is crucial for recog-
nizing the etiology of the adjacent disease, choosing
the ideal treatment, and reducing the worsening of the
patient’s health status (Hallifax et al., 2017). Pleural
effusion is a fluid accumulation in the space between
the parietal and visceral pleura. Various diseases
and conditions, including infections, neoplasms, heart
failure, and chest trauma, can cause pleural effusion.
According to the literature, the late pleural effusion
diagnosis may be associated with worsening the pa-
tient’s health status or lead to other complications,
such as respiratory failure, sepsis, and even death
(Aboudara and Maldonado, 2019).
Chest radiography is an easily accessible tool to
a
https://orcid.org/0000-0002-1102-7722
b
https://orcid.org/0000-0003-3859-6199
c
https://orcid.org/0000-0002-6488-7654
d
https://orcid.org/0000-0001-5080-7660
e
https://orcid.org/0000-0002-9550-5284
detect pleural effusion. However, this finding can
be challenging in cases of small fluid volume, as the
exam can detect volumes above 200 milliliters (Jany
and Welte, 2019). In this way, machine learning can
potentially contribute to early pleural effusion detec-
tion on chest X-rays. In addition, machine learn-
ing models can analyze large chest X-rays datasets to
identify patterns and characteristics associated with
the presence of chest X-ray findings (Bustos et al.,
2020; Liz et al., 2023). Therefore, it is possible
to develop automated classification models that can
identify cases of pleural effusion, generating a double
reading tool.
One of the main challenges in developing ma-
chine learning models to detect chest X-ray findings is
the availability of previously labeled datasets (Cohen
et al., 2020). This scenario involves the arduous pro-
cess of identifying and annotating patient chest im-
ages by experienced radiologists (Bustos et al., 2020).
This absence of labeled data can limit the effective-
ness of machine learning models. Over the past few
years, works in the literature have relied extensively
Zeiser, F., Santos, I., Bohn, H., da Costa, C., Ramos, G., Righi, R., Maier, A., Andrade, J. and Bacelar, A.
Pleural Effusion Classification on Chest X-Ray Images with Contrastive Learning.
DOI: 10.5220/0012205900003584
In Proceedings of the 19th International Conference on Web Information Systems and Technologies (WEBIST 2023), pages 399-405
ISBN: 978-989-758-672-9; ISSN: 2184-3252
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
399

on pre-trained models on natural image datasets, such
as for the ImageNet dataset (Baltruschat et al., 2019;
Zeiser et al., 2021; Guan et al., 2021). However, re-
cent studies have demonstrated problems in capturing
specific medical domain representations using pre-
trained weights (Azizi et al., 2021).
Recent advances address the use of self-
supervision methods to reduce model dependencies
on large datasets (Chen et al., 2020; Azizi et al., 2021;
Zhang et al., 2022). Generally, the work methodol-
ogy is based on a pre-training phase on weakly la-
beled datasets, and then a fine-tuning of the weights
is performed on a fully labeled dataset to improve the
model’s capabilities. However, these methodologies
only have the ability to predict explicitly annotated
findings in the dataset, which still does not solve the
need to perform large-scale manual annotations for
less specific findings.
The availability of chest X-ray images and re-
ports represents a rich source of information for ma-
chine learning models. However, these data cannot be
used directly in supervised learning models without
the production of explicit labels by radiologists. In
this context, this article presents a contrastive learn-
ing model to classify chest X-ray images without ex-
plicit annotations during the training phase. The main
objective is to explore the contrastive learning effec-
tiveness in detecting pleural effusion in chest X-rays
in a context with the scarcity of labeled datasets. Our
method uses contrastive learning to identify key fea-
tures associated with pleural effusion. In addition,
this method can capture more complex characteris-
tics by analyzing natural language present in medi-
cal reports and learning through natural supervision.
Adopting contrastive learning provides a more effi-
cient approach and reduces development costs since
it eliminates the need for extensive data labeling. The
results of our approach demonstrate that contrastive
learning can lead to more accurate and robust classi-
fication models, even when the labeled dataset is lim-
ited or incomplete (Radford et al., 2021). Our main
contributions are listed below:
• We propose a neural network model for weakly
supervised pleural effusion classification on chest
X-ray images. The model can identify and asso-
ciate the images and reports characteristics, opti-
mizing the similarity of the characteristics vector
from two encoders based on Transformers.
• Our experiments demonstrate consistent perfor-
mance improvements in pleural effusion detec-
tion. Furthermore, the results outperform meth-
ods based on supervised learning, suggesting that
explicit labels are not necessary for pleural effu-
sion detection in chest X-ray images.
The remainder of this paper is organized as fol-
lows. Section 2 presents the most significant related
works to define the present study. Next, section 3
presents the methodology of the work. The section 4
details the results and discussion. Finally, Section 5
presents the conclusions of the work.
2 RELATED WORK
The detection of findings in chest X-rays plays a cru-
cial role in the diagnosis and treatment of various dis-
eases. However, this task heavily relies on the ex-
pertise and experience of radiologists, making it sus-
ceptible to errors and human limitations. In recent
years, significant progress has been made in the field
of artificial intelligence, particularly in the develop-
ment of deep learning algorithms for medical image
analysis. Supervised models have been extensively
explored to aid in the analysis of chest X-ray images,
with several studies focusing on this area (Alghamdi
et al., 2021; Bustos et al., 2020; Elgendi et al., 2021;
Han et al., 2021; Rajpurkar et al., 2017; Wang et al.,
2017). These models have shown promising results in
assisting radiologists and improving the accuracy and
efficiency of chest X-ray interpretation.
Supervised learning approaches for classification
based on radiological findings dependent on large
data sets are frequent in the current literature. For
example, (Rajpurkar et al., 2017) developed a ma-
chine learning model that achieved similar accuracy
to a suite of radiologists using a labeled dataset of
over 100,000 images. (Cohen et al., 2020) investi-
gates the performance of models in different domains
and the agreement between them in the X-ray images.
The authors perform experiments to analyze concept
similarity, using a regularization approach in a net-
work to group tasks on different datasets, and observe
the variation between tasks. One of the alternatives
to increase the number of samples for training is data
augmentation (Elgendi et al., 2021). However, data
augmentations commonly applied to tasks in the nat-
ural imagery domain cannot be fully extended to the
health domain. Therefore, labeled datasets are essen-
tial for training machine learning models for X-ray
images (Goceri, 2023).
Obtaining labeled datasets can be complex and la-
borious, especially in resource-poor contexts (Bustos
et al., 2020). In addition, the scarcity of labeled data
can lead to less accurate models and impair the effec-
tiveness of detecting radiological findings in the eval-
uated exams (Alghamdi et al., 2021). Some recent
work has explored the use of unlabeled or partially
labeled datasets to improve the detection of consol-
WEBIST 2023 - 19th International Conference on Web Information Systems and Technologies
400

idation on chest X-ray images. For example, (Wang
et al., 2017) used semi-supervised learning to improve
exam consolidation detection.
Another promising approach is contrastive learn-
ing, which can be applied to unlabeled or partially
labeled datasets (Zhang et al., 2022). This feature
uses the similarity and differences between pairs of
images to learn discriminative characteristics (Chen
et al., 2020). Therefore, the method is beneficial when
small-scale labeled data is available. For example,
(Han et al., 2021) used contrastive learning in con-
junction with transfer learning to improve consolida-
tion detection on chest X-rays.
However, works in the current literature are pri-
marily based on fully labeled datasets or require par-
tially labeled datasets in the training process. There-
fore, this work proposes to investigate the effective-
ness of contrastive learning to improve the detection
of pleural effusion in chest X-rays in the context of
training models with weak-labeled datasets.
3 MATERIALS AND METHODS
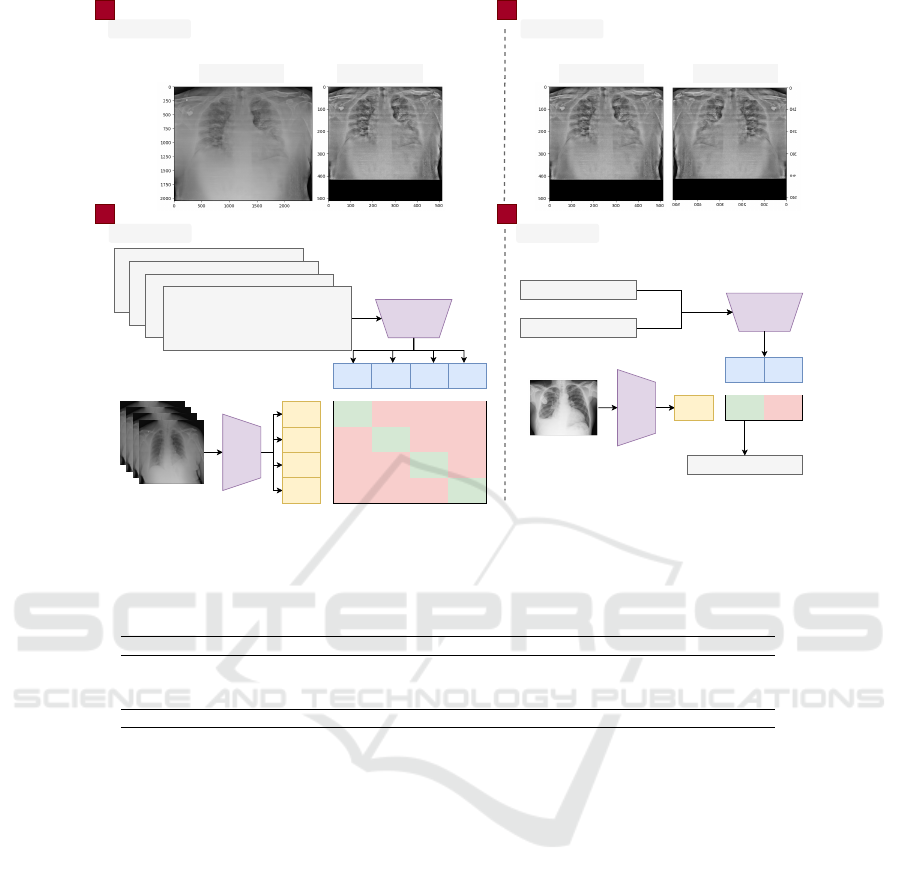
An overview of the methodology used in this work
is presented in Fig. 1. Our methodology can be di-
vided into four main steps: pre-processing, data aug-
mentation, training, and testing. Pre-processing con-
sists of resizing the image, normalizing the contrast,
and processing the report (Section 3.2). The data aug-
mentation step describes the methods used to generate
synthetic images (Section 3.3). Finally, in the train-
ing stage, the models and parameters used are defined
(Section 3.4).
3.1 Dataset
We used three datasets to develop the model. For the
training step, we retrospective collected data from pa-
tients during the COVID-19 pandemic from two hos-
pitals in Porto Alegre/RS. The information collected
comprises clinical data, X-ray images and reports of
hospitalizations from 2020 to 2022. The ethics com-
mittee of each hospital approved the present study un-
der the Certificate of Submission for Ethical Consid-
eration (CAAE number 33540520.6.3004.5327) . In
addition, this document follows the General Data Pro-
tection Law (LGPD) recommendations. In Tab. 1, the
information relating to Private datasets 01 and 02 is
provided.
We used the PADCHEST public dataset for the
testing step. PADCHEST consists of chest X-rays
with reports. The dataset includes more than 160,000
images of 67,000 patients that were interpreted and
reported by radiologists at Hospital San Juan (Spain)
from 2009 to 2017, encompassing six views of dif-
ferent incidences, additional information on image
acquisition, and patient demographics. The dataset
comprises 19 differential diagnoses, with 27% of re-
ports manually annotated by physicians. In this sense,
we used only manually annotated X-ray images. In
Tab. 1, we present the information for each dataset.
3.2 Pre-Processing
We processed the images from the three datasets using
the same methodology. Given the tenuous differences
between healthy tissues and those affected by any al-
teration, contrast enhancement is commonly used in
the literature (Diniz et al., 2018). Furthermore, con-
trast enhancement can help increase the performance
of deep learning architectures (Pooch et al., 2020).
Therefore, we applied the Contrast-Limited Adaptive
Histogram Equalization (CLAHE) in the chest X-ray
images. CLAHE subdivides the image into subareas
using interpolation between edges. To avoid noise
build-up, use a gray threshold level, redistributing
pixels above that threshold in the image. The CLAHE
can be defined by:
p = [p
max
− p
min
] ∗ G( f ) + p
min
(1)
where p is the pixel’s new gray level value, the values
p
max
and p
min
are the pixels with the lowest and high-
est values in the neighborhood, and G( f ) corresponds
to the cumulative distribution function (Zuiderveld,
1994).
Human anatomy is unique to each individual. This
aspect is reflected in the different anatomical sizes of
patients’ chests. In this way, the X-ray images have
different sizes. However, to process the X-ray images
in deep learning models, the images need a standard
width and height. Therefore, we resized the images
to 512X512 pixels. The reduction was proportional
to the width and height, with a zero padding for the
axis with the smallest size. The pre-trained architec-
ture delimited the image resolution, which will be dis-
cussed in Section 3.4. In Fig. 2, we show an example
with the original and preprocessed X-ray images. We
can observe (Fig. 2) that the image suffered a down-
sampling changing from approximately 2000X 3000
pixels to 512X512 pixels. In addition, the area related
to the lungs is highlighted about the other structures
represented in the image.
Finally, we pre-processed the Private datasets 01
and 02 X-ray reports. We keep only the alpha charac-
ters, removing special characters, numbers, and punc-
tuation from the reports. For the PADCHEST dataset,
it was only necessary to translate the pleural effusion
Pleural Effusion Classification on Chest X-Ray Images with Contrastive Learning
401
Text Encoder
Image
Encoder
I1
I2
I3
I4
T1
T2
T3
T4
I1 * T1
I1 * T2
I1 * T3
I1 * T4
I2 * T1
I2 * T2
I2 * T3
I2 * T4
I3 * T1
I3 * T2
I3 * T3
I3 * T4
I4 * T1
I4 * T2
I4 * T3
I4 * T4
Training
tubo endotraqueal com extremidade no bronquio
principal direito cateter venoso central direita com
extremidade na projecao da veia cava superior demais
aspectos sem modificacao em relacao ao exame
anterior da vespera
faixa atelectasica em terco medio inferior do pulmao
direito infiltrado reticular difuso bilateral elevacao de
hemicupula diafragmatica direita aumento da area
cardiaca aorta ateromatosa seios costofrenicos laterais
livres
area de reducao da transparencia do parenquima
pulmonar na metade inferior do pulmao esquerdo
podendo representar area de consolidacao com
obliteracao do recesso costofrenico relacionada
moderado derrame pleural mediastino centrado indice
cardiotoracico dentro da normalidade recesso
costofrenico direito livre
presenca de volumoso derrame pleural esquerda com
discreto desvio do mediastino para direita nao ha
evidencia de lesao consolidativa ou tumescente
grosseira em pulmao direito nao foi possivel
determinar area cardiaca nao ha evidencia de derrame
pleural no seio costofrenico lateral direito
Testing
I1
Text Encoder
Image
Encoder
T1
T2
I1 * T1
I1 * T2
sem presenca derrame pleural
presenca derrame pleural
presenca derrame pleural
Pre-processing
Original X-ray Pre-processed X-ray
Data Augmentation
Pre-processed X-ray Flipped X-ray
1
STEP
2
STEP
3
STEP
4
STEP
Figure 1: Summary of the proposed methodology. The text and image encoders are trained together to predict the correct pairs
from a training batch. The text encoder is fed with generic text, and the image encoder with the chest X-ray for prediction.
Table 1: Chest X-ray datasets for training and testing the architecture. Total.: Total; Image: Image; Man.: Manual; and Aut.:
Automatic.
Dataset Patients Annotation Label Tot. Img. Img. Used Used in
Private 01 697 Man. Report 6.650 6.650 Training
Private 02 1.754 Man. Report 7.084 7.084 Training
PADCHEST 69.882 Man./Aut. Report and Findings 160.000 17.513 Teste
Total 72.333 - Report and Findings - 173.734 -
finding from Spanish into Portuguese, which was per-
formed by automatic substitution.
3.3 Data Augmentation
We used a data augmentation methodology to in-
crease the cases in the training set. In this context, we
applied a horizontal flip to each training set image.
The choice of only one data augmentation method
is related to the image characteristics. X-ray exams
represent anatomical structures that can be missed or
distorted by other data augmentation methods such as
distortion, shearing, or cropping.
3.4 Contrastive Learning Architecture
Current models used in the field of computer vi-
sion require large datasets labeled (Dosovitskiy et al.,
2020). Specifically, in the context of imaging stud-
ies, obtaining large labeled datasets is a significant
challenge. In this way, methods that can learn from
already established information may be able to scale
with greater generalization for the day-to-day use
of radiologists. In this sense, recent advances in
contrastive learning methods present a possibility of
learning representations in images using natural lan-
guage (Radford et al., 2021; Zhang et al., 2022).
In this way, our architecture shown in Fig. 1 is
based on Connecting text and images (CLIP) (Rad-
ford et al., 2021). The CLIP architecture is based on
the idea of zero-shot learning, seeking to find relation-
ships between image and text pairs. This association
is built using two encoders, one responsible for ex-
tracting features representing the image and another
for the text. As a result, the model’s objective func-
tion is that both encoders produce feature vectors for
similar images and texts.
3.4.1 Training
The parameters for the encoders followed the CLIP
architecture (Radford et al., 2021). The image en-
coder is based on the ViT-B/32 architecture pre-
WEBIST 2023 - 19th International Conference on Web Information Systems and Technologies
402
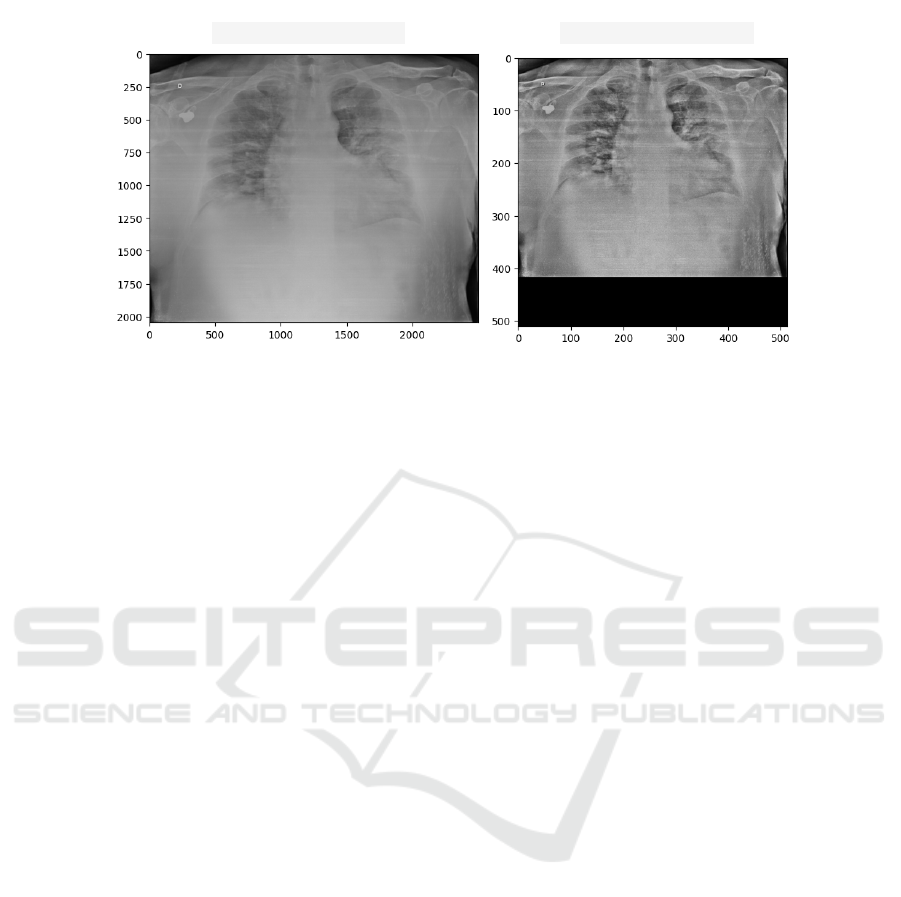
(a) Original X-ray (b) Pre-processed X-ray
Figure 2: Example of an original chest X-ray image (a) and pre-processed (b).
trained with CLIP model weights. A transformer ar-
chitecture with 12 layers of size 512 and 8 attention
heads was developed for the text encoder. The maxi-
mum length of the text string was 76 characters. The
implementation of the deep learning model followed
the hyper-parameters of the CLIP model.
The model was trained for 100 epochs using the
Adam optimizer with an adaptive learning rate of
0.00001. The batch size was 32 images. The model
was trained with the loss function of cosine similarity,
maximizing the similarity between the correct image
and text pairs.
3.4.2 Evaluation
For model evaluation, each image is processed with
two synthetic texts in Portuguese: (a) presence of
pleural effusion; and (b) without presence of pleural
effusion. First, the respective encoders calculate the
feature vector for the image and a feature vector for
each synthetic text. Then, the cosine similarity is cal-
culated with the normalization of the result performed
using a softmax function. The output of the softmax
function then represents the binary classification for
the presence or absence of pleural effusion on the
chest X-ray. We used the area under the receiver oper-
ating characteristic (AUC), precision/recall curve, ac-
curacy, sensitivity, specificity, and F1-score as perfor-
mance metrics for evaluating the model. In addition,
we performed a bootstrap to generate the performance
metrics confidence intervals.
4 RESULT AND DISCUSSION
This section presents the results of the proposed
method and the comparison with the current literature
for the detection of pleural effusion. The best weights
were chosen automatically based on the validation set
error. Tab. 2 presents the performance obtained for
the evaluation metrics in the PADCHEST dataset and
the respective comparison with the current literature.
Few studies in the current literature specifically
focus on the finding of pleural effusion. Therefore,
direct comparison and identifying what could be con-
sidered state o the art for this specific finding is com-
plex. Therefore, the survey of related works was
mainly based on work that detected different findings
in chest X-ray images.
Despite the different objectives, the results pre-
sented in Tab. 2 show that even without labels during
training, the methodology based on the CLIP (Rad-
ford et al., 2021) architecture is capable of achiev-
ing results very close or superior to supervised learn-
ing methods. At this point, it is essential to highlight
the sensitivity (88.54%) for detecting pleural effusion,
demonstrating that the model is capable of identifying
the findings.
Regarding the ROC curve (Fig. 3), the proposed
method obtained a value of 0.900. Compared with
other classification methods using the same dataset
(Liz et al., 2023), our approach achieved superior per-
formance in terms of AUC-ROC. Furthermore, the
method proposed by (Liz et al., 2023) used part of the
PADCHEST dataset in the training process. While in
our method, PADCHEST was used only for method
testing. Therefore, it is possible that our method has
a superior ability to generalize multi-center findings
and may be more susceptible to use in clinical settings
to aid in diagnosing pleural effusion.
The results of our study demonstrate that the con-
trastive learning method can be a practical approach
to classifying pleural effusions on X-ray images. Al-
though there is space for improvement in the classifi-
cation performance metrics and extension for a radio-
Pleural Effusion Classification on Chest X-Ray Images with Contrastive Learning
403

Table 2: Comparison with related works. AUC: area under the receiver operating characteristic. Acc: Accuracy. Sen:
Sensibility. Spe: Specificity. F1: F1-score.
Study Dataset AUC Acc Sen Spe F1
(Cohen et al., 2020) Multiple 0.890 - - - -
(Guan et al., 2021) NIH ChestX-ray14 0.835 - - - -
(Ibrahim et al., 2022) NIH ChestX-ray14 - 88.86 - - -
(Liz et al., 2023) PADCHEST 0.656 - - - 56.70
(Rajpurkar et al., 2017) NIH ChestX-ray14 0.863 - - - -
(Serte and Serener, 2021) ChestX-ray8 0.780 75.00 100.00 67.00 -
(Zaidi et al., 2021) NIH ChestX-ray14 0.874 98.20 - - -
Ours PADCHEST 0.900 86.28 88.54 76.91 68.41
ROC Curve
True Positive Rate
False Positive Rate
Figure 3: ROC curve for the PADCHEST dataset.
logical multi-finding classification model, the current
results are promising and open possibilities for stud-
ies of our model in clinical settings.
5 CONCLUSION
In conclusion, this study explored the use of con-
trastive learning to classify pleural effusion on chest
X-rays. The results showed that the proposed
approach could outperform reference models and
achieve significantly higher accuracy. In addition, the
use of contrastive learning can be helpful in scenarios
with restricted information about the clinical behav-
ior of the disease, such as outbreaks and pandemics
of lesser-known or emerging diseases.
This work has a few limitations, as described next.
The first is related to the technical limitation of be-
ing a model capable of finding only one finding per
image. Thus, studying methods that can identify dif-
ferent image findings is a prospect for future work.
Another limiting aspect is the data set for training.
In this way, using sets with a greater variety of find-
ings is foreseen, allowing more excellent reliability
for clinical application. Finally, interpretability stud-
ies identify how the model creates the representations
and how it relates to them during the prediction pro-
cess. These studies can be extended to propose inter-
pretability methods for radiologists.
The benefits of using contrastive learning to accel-
erate the creation of business models for real use are
significant. The proposed method can help save time
and valuable resources in creating machine learning
models for healthcare applications, enabling faster
and more accurate diagnoses, and improving the qual-
ity of the patient’s hospital course. These advantages
make the proposed approach a promising tool to en-
hance the analysis of chest X-ray images in real clin-
ical scenarios.
ACKNOWLEDGEMENTS
The authors would like to thank the Coordination for
the Improvement of Higher Education Personnel -
CAPES (Financial Code 001), the National Council
for Scientific and Technological Development - CNPq
(Grant numbers 309537/2020-7 e 40572/2021-9), and
the Research Support Foundation of the State of Rio
Grande do Sul - FAPERGS (Grant number 21/2551-
0000118-6) for your support in this work.
REFERENCES
Aboudara, M. and Maldonado, F. (2019). Update in the
management of pleural effusions. Medical Clinics,
103(3):475–485.
Alghamdi, H. S., Amoudi, G., Elhag, S., Saeedi, K., and
Nasser, J. (2021). Deep learning approaches for de-
tecting covid-19 from chest x-ray images: A survey.
Ieee Access, 9:20235–20254.
Azizi, S., Mustafa, B., Ryan, F., Beaver, Z., Freyberg, J.,
Deaton, J., Loh, A., Karthikesalingam, A., Kornblith,
WEBIST 2023 - 19th International Conference on Web Information Systems and Technologies
404

S., Chen, T., et al. (2021). Big self-supervised models
advance medical image classification. In Proceedings
of the IEEE/CVF International Conference on Com-
puter Vision, pages 3478–3488.
Baltruschat, I. M., Nickisch, H., Grass, M., Knopp, T., and
Saalbach, A. (2019). Comparison of deep learning
approaches for multi-label chest x-ray classification.
Scientific reports, 9(1):1–10.
Bustos, A., Pertusa, A., Salinas, J.-M., and de la Iglesia-
Vay
´
a, M. (2020). Padchest: A large chest x-ray image
dataset with multi-label annotated reports. Medical
image analysis, 66:101797.
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020).
A simple framework for contrastive learning of visual
representations. In International conference on ma-
chine learning, pages 1597–1607. PMLR.
Cohen, J. P., Hashir, M., Brooks, R., and Bertrand, H.
(2020). On the limits of cross-domain generalization
in automated x-ray prediction. In Medical Imaging
with Deep Learning, pages 136–155. PMLR.
Diniz, J. O. B., Diniz, P. H. B., Valente, T. L. A., Silva,
A. C., de Paiva, A. C., and Gattass, M. (2018). De-
tection of mass regions in mammograms by bilateral
analysis adapted to breast density using similarity in-
dexes and convolutional neural networks. Computer
methods and programs in biomedicine, 156:191–207.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Elgendi, M., Nasir, M. U., Tang, Q., Smith, D., Grenier,
J.-P., Batte, C., Spieler, B., Leslie, W. D., Menon,
C., Fletcher, R. R., et al. (2021). The effectiveness
of image augmentation in deep learning networks for
detecting covid-19: A geometric transformation per-
spective. Frontiers in Medicine, 8:629134.
Goceri, E. (2023). Medical image data augmentation: tech-
niques, comparisons and interpretations. Artificial In-
telligence Review, pages 1–45.
Guan, Q., Huang, Y., Luo, Y., Liu, P., Xu, M., and Yang, Y.
(2021). Discriminative feature learning for thorax dis-
ease classification in chest x-ray images. IEEE Trans-
actions on Image Processing, 30:2476–2487.
Hallifax, R., Talwar, A., Wrightson, J., Edey, A., and Glee-
son, F. (2017). State-of-the-art: Radiological inves-
tigation of pleural disease. Respiratory medicine,
124:88–99.
Han, Y., Chen, C., Tewfik, A., Ding, Y., and Peng, Y.
(2021). Pneumonia detection on chest x-ray using
radiomic features and contrastive learning. In 2021
IEEE 18th International Symposium on Biomedical
Imaging (ISBI), pages 247–251. IEEE.
Ibrahim, R. F., Yhiea, N., Mohammed, A. M., and Mo-
hamed, A. M. (2022). Pleural effusion detection us-
ing machine learning and deep learning based on com-
puter vision. In Proceedings of the 8th International
Conference on Advanced Intelligent Systems and In-
formatics 2022, pages 199–210. Springer.
Jany, B. and Welte, T. (2019). Pleural effusion in
adults—etiology, diagnosis, and treatment. Deutsches
¨
Arzteblatt International, 116(21):377.
Liz, H., Huertas-Tato, J., S
´
anchez-Monta
˜
n
´
es, M., Del Ser,
J., and Camacho, D. (2023). Deep learning for under-
standing multilabel imbalanced chest x-ray datasets.
Future Generation Computer Systems.
Pooch, E. H. P., Alva, T. A. P., and Becker, C. D. L. (2020).
A deep learning approach for pulmonary lesion identi-
fication in chest radiographs. In Brazilian Conference
on Intelligent Systems, pages 197–211. Springer.
Radford, A., Kim, J. W., Hallacy, C., Ramesh, A., Goh, G.,
Agarwal, S., Sastry, G., Askell, A., Mishkin, P., Clark,
J., et al. (2021). Learning transferable visual models
from natural language supervision. In International
conference on machine learning, pages 8748–8763.
PMLR.
Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan,
T., Ding, D., Bagul, A., Langlotz, C., Shpanskaya, K.,
et al. (2017). Chexnet: Radiologist-level pneumonia
detection on chest x-rays with deep learning. arXiv
preprint arXiv:1711.05225.
Serte, S. and Serener, A. (2021). Classification of covid-19
and pleural effusion on chest radiographs using cnn
fusion. In 2021 International Conference on INno-
vations in Intelligent SysTems and Applications (IN-
ISTA), pages 1–6. IEEE.
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Sum-
mers, R. M. (2017). Chestx-ray8: Hospital-scale chest
x-ray database and benchmarks on weakly-supervised
classification and localization of common thorax dis-
eases. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 2097–
2106.
Zaidi, S. Z. Y., Akram, M. U., Jameel, A., and Al-
ghamdi, N. S. (2021). Lung segmentation-based pul-
monary disease classification using deep neural net-
works. IEEE Access, 9:125202–125214.
Zeiser, F. A., Costa, C. A. d., Ramos, G. d. O., Bohn, H.,
Santos, I., and Righi, R. d. R. (2021). Evaluation of
convolutional neural networks for covid-19 classifi-
cation on chest x-rays. In Intelligent Systems: 10th
Brazilian Conference, BRACIS 2021, Virtual Event,
November 29–December 3, 2021, Proceedings, Part
II 10, pages 121–132. Springer.
Zhang, Y., Jiang, H., Miura, Y., Manning, C. D., and Lan-
glotz, C. P. (2022). Contrastive learning of medical
visual representations from paired images and text. In
Machine Learning for Healthcare Conference, pages
2–25. PMLR.
Zuiderveld, K. (1994). Graphics gems iv. In Heckbert,
P. S., editor, Graphics Gems, chapter Contrast Lim-
ited Adaptive Histogram Equalization, pages 474–
485. Academic Press Professional, Inc., San Diego,
CA, USA.
Pleural Effusion Classification on Chest X-Ray Images with Contrastive Learning
405
