
Pill Metrics Learning with Multihead Attention
Rich
´
ard R
´
adli
a
, Zsolt V
¨
or
¨
osh
´
azi
b
and L
´
aszl
´
o Cz
´
uni
c
University of Pannonia, 8200 Veszpr
´
em Egyetem u. 10., Hungary
Keywords:
Metrics Learning, Pill Recognition, Self-Attention, Multihead Attention, Multi-Stream Network, Siamese
Network, YOLO.
Abstract:
In object recognition, especially, when new classes can easily appear during the application, few-shot learning
has great importance. Metrics learning is an important elementary technique for few-shot object recognition
which can be applied successfully for pill recognition. To enforce the exploitation of different object features
we use multi-stream metrics learning networks for pill recognition in our article. We investigate the usage of
multihead attention layers at different parts of the network. The performance is analyzed on two datasets with
superior results to a state-of-the-art multi-stream pill recognition network.
1 INTRODUCTION
It is claimed that drug errors are the most frequent
mistakes in healthcare (Cronenwett et al., 2007). Pill
recognition systems can have a great positive impact
on the quality of pill dispensing considering either
home usage or automatic pill selection in large scale
systems.
The different problems, originating from
taking the wrong medications, can be so serious
worldwide that the WHO has chosen Medication
Safety as the theme for the World Patient Safety
Day in 2022 (https://www.who.int/campaigns/
world-patient-safety-day/2022). In theory, pills are
designed to have discriminating several features
(size, color, shape, engravings, imprints, etc.),
unfortunately, there are many factors increasing the
uncertainty of recognition:
• pill photographs are taken under various
conditions (e.g. illumination, viewing angle,
object distance, camera settings, backgrounds);
• since pills have small size, local features are often
not visible or distorted on the images.
Beside these influences the number of possible pill
classes is very large (it can reach up over 10000) and
few-shot learning is required in many applications
since often new pills should be added to a system after
the standard training process.
a
https://orcid.org/0009-0009-3160-1275
b
https://orcid.org/0009-0004-3032-8784
c
https://orcid.org/0000-0001-7667-9513
To foster the development of reliable solutions the
United States National Library of Medicine (NLM)
announced an algorithm challenge on pill recognition
in 2016 (Yaniv et al., 2016). After the evaluation
of results, the accuracy of the top three submissions
seemed not to be sufficient for the development
of a mobile online service for matching consumer
quality images. In our article we make several steps
to improve the winner model architecture of the
competition ( (Zeng et al., 2017)) and its descendant
(Ling et al., 2020). While our proposed method is still
a multi-stream network for image embedding, trained
to differentiate pill classes by their images, there are
several important differences. These differences are
detailed in Section 2 and 4.
In our article we report two-sided pill tests. This
means that each side of a pill belongs to the same class
in contrast to one-side tests where each side of a pill
is in different classes. The main contributions of our
paper:
• introduction of a new solution of pill recognition
with SOTA results in two-sided tests;
• comparison of two backbones;
• comparison of different alternatives for the
inclusion of attention layers.
The proposed methods were tested on two different
data sets: CURE and OGYEI (see details in
Section 3).
We refer to our model as multi-stream
self-attention (MS-SA), and multi-stream multihead
attention (MS-MHA) in our article.
132
Rádli, R., Vörösházi, Z. and Czúni, L.
Pill Metrics Learning with Multihead Attention.
DOI: 10.5220/0012235500003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 1: KDIR, pages 132-140
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

2 A BRIEF OVERVIEW OF
RELATED ARTICLES
The pill recognition problem is very similar to other
recognition tasks, such as the recognition of stamps,
coins, or other small objects with large number of
classes. Due to the limited size of our paper we
give a short overview of such techniques which were
designed for and tested on drug datasets.
The winner of the aforementioned tablet
recognition competition (Zeng et al., 2017) used a
multi-stream technique in which separate teaching
CNNs processed the color, gray, and gradient images
of already localized pills. A knowledge distillation
model compression framework then condensed the
training CNNs into smaller footprint CNNs (student
CNNs), employed during inference time. CNNs
were designed to embed features in a metric space,
where cosine distance is utilized as the metric
to determine how similar the features produced
by CNNs are to each other. During the training
of the streams Siamese networks were used with
three inputs: the anchor image, a positive, and a
negative sample, while the applied triplet loss was
responsible to minimize the distance between the
anchor and positive samples, and to increase the
distance between the anchor and negative samples.
This model was improved in (Ling et al., 2020)
with better accuracy proven by several tests on the
CURE dataset. They omitted the teacher-student
compression approach, but added a separate OCR
(optical character recognition) stream, and a fusion
network to process the output of the streams. The
OCR stream carried out text localization, geometric
normalization, and solved the generation of feature
vectors with a deep text recognizer called Deep
TextSpotter (Busta et al., 2017). Beside the OCR
stream RGB, texture, and contour streams were used;
the segmentation, to generate inputs for the streams,
was achieved with an improved U-Net model. We
followed a similar approach to (Ling et al., 2020)
but we made several modifications. The OCR
method was replaced with LBP (local binary pattern)
(Ojala et al., 1994) streams, we applied attention
mechanisms, used different localization, and we
worked with several versions of EfficientNet instead
of custom DNN backbones. Details and justification
is given in Section 4.
Generic object detectors, in their native form,
don’t fit perfectly the pill recognition problem due
to the few-shot learning tasks in many use-case
scenarios. However, the generic methods have made
great improvements in recent years, so we have
included some interesting approaches in our brief
review.
In (Tan et al., 2021) three object detectors
(YOLOv3, RetinaNet, and SSD) were compared on
a custom dataset, resulting only in small differences
in mAP - mean of average precision (∼ 2%, all
above 0.80). We also provide a comparison of our
multi-stream solution with YOLOv7 (Wang et al.,
2023) in Section 5.2.
In (Nguyen et al., 2022) a deep learning-based
approach was proposed to solve the contextual
pill recognition problem. The solution used a
prescription-based knowledge graph, representing the
relationship between pills. A graph embedding
network extracts pills’ relational features and a
framework is applied to fuse the graph-based
relational information with the image-based visual
features for the final classification. The drawback of
this method is that it requires medical prescriptions, or
equivalently it can be applied when there are multiple
pills on a image.
In (Heo et al., 2023) the authors trained not only
RGB images of the pills but also imprinted characters.
In the pill recognition step, the different modules
separately recognize both the features of pills and
their imprints, while it can correct the recognized
imprint to fit the actual data of other features. A
trained language model was applied in the imprint
correction. It was shown through an ablation study
that the language model could significantly improve
the pill identification ability of the system.
In contrast to these approaches, in our solution
we have avoided the use of specific language
models, otherwise the training would require the
input and processing of textual information and/or
language-specific OCR modules.
3 DATASETS
There are several image datasets for pill
recognition (Ling et al., 2020), (VAIPE, 2008), (Yaniv
et al., 2016). For one part of our experiments we have
chosen CURE image database (Ling et al., 2020)
since it has many different backgrounds, varying
illumination conditions, and accuracy values are
available for the model of (Ling et al., 2020). In
CURE, images are divided into reference quality
and consumer grade images: typically the former
is utilized for positive and negative examples in the
triplets. The disadvantage is that reference images
are synthetically generated from consumer images,
artificially replacing their backgrounds.
To simulate the operation of an automatic
dispensing utility, we needed a data set with
Pill Metrics Learning with Multihead Attention
133

well-defined conditions: a fixed scale (since size
can be important information for recognition),
a homogeneous background, and two different
illumination settings: a diffuse light source from
above and alternatively a linear light from a lower
elevation on the side to make engravings more visible.
These requirements are satisfied in our custom
OGYEI data set which can be utilized in use-cases for
pill recognition under controlled environments. We
summarize the main parameters of the two datasets in
Table 1.
Table 1: Comparison of CURE and OGYEI datasets.
CURE OGYEI
Number of
pill classes
196 78
Number of
images
8973 3154
Image
resolution
800×800
2448×2448
2465×1683
Instance
per class
40-50 40-60
Segmentation
labels
no fully
Backgrounds 6 1
Imprinted
text labels
Yes Yes
Reference/
customer images
Both
Reference
quality images
Figure 1 shows sample images of two pills from
the OGYEI reference-quality dataset under different
lighting conditions. (Please note that all attempts on
the recognition experiments in our paper were based
on a single image, so the use of lighting information
is a task for future work.)
4 MULTI-STREAM METRICS
LEARNING
The general overview our the proposed model
is in Figure 2 (more precisely the EfficientNet
model based alternatives are named EffNetV1+SA
and EffNetV2+SA in Section 5). Since metrics
embedding does not solve the problem of object
localization, the bounding box of pills are determined
first. Then four streams with very similar structures
perform the first part of image embedding. In the
second phase, the information of the branches is fused
to obtain the final tablet representation.
Figure 1: Example images of two pills (Milurit and
Dulodet-60) from the OGYEI dataset illuminated from the
side (left), and from above (right).
Figure 2: General overview of our multi-stream model.
4.1 Localization of Pills
In theory, two types of errors can occur when
detecting and recognizing a single, or multiple pills
on an image: either the tablet is not found, or
other objects are labeled as pills and the correctly
detected pill is misclassified. In case of single stage
detectors the training of the whole process (detection,
localization, and recognition) is combined in a single
step, but metrics learning only solves the recognition
part.
Unfortunately, many papers does not focus on
the localization of pills on the input images. For
example (Tan et al., 2021) uses only pill images with
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
134

dark backgrounds. The authors of (Nguyen et al.,
2022) use their own dataset with tightly cropped pill
images and explicitly state that object localization is
out of the scope of their paper. Also article (Heo
et al., 2023) does not deal with it, assuming that
images contain single pill in front of homogeneous
background.
Contrary, in paper (Ling et al., 2020) great effort
is payed to find the regions of pills placed over
various backgrounds. Even a modified U-Net model
was developed for the segmentation of pill areas.
Unfortunately, their improved U-Net model was not
available for us and we could not reproduce the
same model from their descriptions. We investigated
the standard U-Net (Ronneberger et al., 2015) but
were not satisfied with its performance. In many
cases it detected noise on various backgrounds which
should have been further processed to exclude false
positive detections. Since exact segment borders are
not required for our multi-stream model we decided
to train YOLOv7 (Wang et al., 2023) for class
detection. (Please note that the U-Net model has 35M
parameters, while the standard YOLOv7 has only
slightly more than 36M parameters.) To illustrate the
problem with noisy detections see Figure 3 and 4 for
an example.
Figure 3: Detected untrained object and its bounding box
from the CURE dataset generated by YOLOv7.
In order to generate the bounding boxes for
multi-stream embedding we trained YOLOv7 on
an augmented dataset of CURE reference subset,
resulting in 105661 training images. The details of
this augmentation process are given in Subsection
4.1.1. Now all pill classes were merged to only one
class (pill object), the network was learning for 5
epochs with default settings. In such cases where
the trained model struggled to find any objects in
the tests, we lowered the confidence threshold from
the original 0.45 to 0.001. Despite these efforts, we
Figure 4: Example for a binary mask of a trained standard
U-Net segmentation model on the input of Figure 3.
still encountered a handful of images where YOLOv7
could not detect any pills, even with the reduced
confidence threshold. In such cases, we made the
decision to pass the whole image to the multi-stream
network as it is.
Evaluation of the detection was carried out on a
separated test set of the CURE dataset (this means
class segregated training and testing). Out of 1716
test instances, the network successfully identified
1628 objects and only failed to find anything on 88
examples. 20 false positive cases have been reported,
thus the testing resulted precision 0.9878 and recall
0.9484. After lowering the threshold, we found 60
more pills that could not be identified with the first
setting.
We also evaluated the performance of YOLOv7
localization (trained only on the augmented CURE
dataset) on the test set of the OGYEI image dataset.
Out of the 474 test images from OGYEI it correctly
detected 464 images, and only discarded 10 images,
having only 1 false positive case, with default settings.
It resulted precision 0.9978 and recall 0.9789.
4.1.1 Augmentation Process
Data augmentation is a crucial technique in deep
learning to enhance the model’s performance and
robustness. It is especially true for pill localization
or recognition, since it is difficult to obtain several
real images of the large number of classes with
various imaging conditions - to avoid the learning
of characteristics other than the pills discriminating
features. In this subsection we explain the data
augmentation processes and techniques we applied
to the images of the CURE dataset for the purpose
of training YOLOv7 as our bounding box detector.
It is important to note, that for the training of our
multi-stream models the original CURE images were
Pill Metrics Learning with Multihead Attention
135
used.
The CURE dataset comprises two main sub-sets
as mentioned in Section 3: the reference set and
the customer set. Our focus was on the reference
images, which are characterized by homogeneous
backgrounds. We split this dataset into train and test
sets, where the classes of the two sets were disjoint.
Before applying any augmentation method the images
(388 pictures) had to be segmented with pixel-wise
accuracy since later we changed their background (in
step 3 given below). Augmentation was carried out in
three consecutive steps:
1. White balance adjustments, Gaussian smoothing,
brightness modifications, rotation, shifting,
zooming, horizontal and vertical flipping.
2. Repeating the above steps to have large number of
combinations of the different effects.
3. Changing the background of the images, utilizing
the DTD (Describable Textures Dataset) dataset
(Cimpoi et al., 2014).
This augmentation process resulted in 105661
images, which we divided into training and testing
parts for the purpose of pill localization (as described
in the previous section).
4.2 Feature Streams
The main idea behind multi-stream processing is to
persuade the sub-networks to focus on different kinds
of features. For this reason different pre-processing
steps are done in the streams:
1. RGB: color images are directly fed to a
CNN for metrics embedding. We evaluated
both EfficientNet-B0 (Tan and Le, 2019)
and EfficientNetv2 S (Tan and Le, 2021).
EfficientNet-B0 has significantly less parameters
than the CNN of (Ling et al., 2020) (5.3 million
vs. 9 million) and it is well-optimized for
similar tasks. EfficientNetv2 is larger (21.4M
parameters) but is reported to be more accurate
in ImageNet tasks and faster in train time. The
same networks were used in all streams but
with smaller number of parameters due to their
grayscale input images.
2. Contour: images are generated by running the
Canny edge detector on smoothed grayscale
version of images (applying a 7 × 7 Gaussian
kernel).
3. Texture: images are created by subtracting the
smoothed and grayscale versions of pill images.
4. Local Binary Patterns (LBP) (Ojala et al.,
1994): LBP is a popular hand crafted local
descriptor for many computer vision tasks
including handwritten or printed OCR (Liu et al.,
2010), (Hassan and Khan, 2015). That is the
reason why we omitted the special OCR stream
of (Ling et al., 2020) but computed the LBP
images of the grayscale inputs and used them in
similar streams as the others.
All streams received the bounding box defined pill
images of resolution 224 ×224 detected by YOLOv7
as described above.
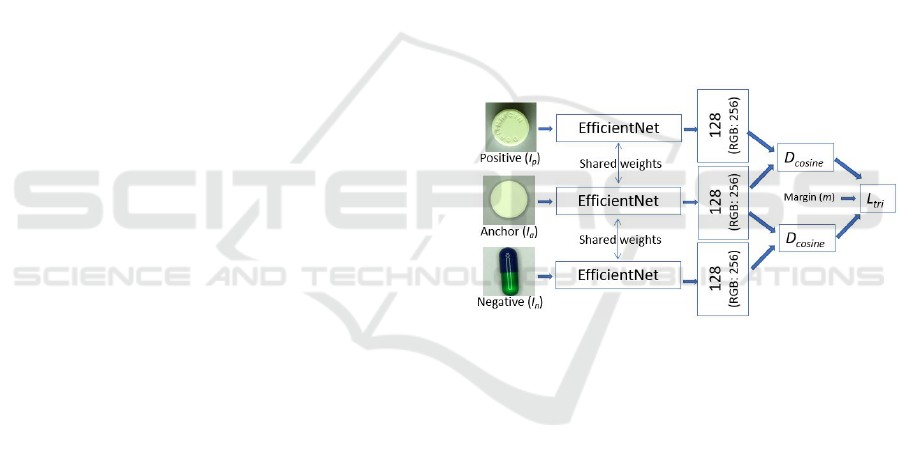
4.3 Training of Streams
For the training of the stream networks Siamese
neural networks with three inputs (anchor - I
a
,
positive example - I
p
, and negative example I
n
) are
used (see illustration in Figure 5). In the model
of Ling et al. (Ling et al., 2020) relatively simple
CNNs were used, in our models we implemented
EfficientNet-B0 and EfficientNetv2 S as already
mentioned in the previous subsection.
Figure 5: The training of streams in Siamese setup.
Each stream is independently trained for metrics
embedding following the batch-all strategy, when any
regular triplet can be used (Ling et al., 2020). During
the training of the high-level fusion network, only
those input triplets are used which were too difficult
to embed in this phase (it is called hard triplet mining,
see Eq. 2).
For loss function L
tri
, in the streams and in the fusion
network, we use the formula introduced in (Schroff
et al., 2015):
L
tri
=
∑
∀(I
a
,I
p
,I
n
)
[m + D(f (I
a
), f (I
p
))
−D( f (I
a
), f (I
n
))]
+
(1)
for all triplets (I
a
, I
p
, I
n
) in a batch, where margin m is
set to 0.5, D is the cosine distance of feature vectors
(generated by network f ), and [x]
+
= max{0, x}.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
136

4.4 Fusion of Streams
Before the concatenation of the embedding vectors
we implemented the attention encoder of the famous
mechanism introduced in (Vaswani et al., 2017) in
each stream. To fuse the information of the streams
we concatenated the output vectors and applied full
connections in one hidden and one output layer to
generate the final embedding (see Figure 2). During
the training of the fusion network streams were frozen
and only the top layers were trained by such triplets
(I
a
, I
p
, I
n
) which satisfied the following criterion:
D( f
hier
(I
a
), f
hier
(I
n
)) −D( f
hier
(I
a
), f
hier
(I
p
)) < m,
(2)
where f
hier
symbolizes the hierarchical model
including the streams and the fusing layers.
4.5 Attention Mechanism Alternatives
Attention and self-attention mechanisms have
been widely used since their appearance (Vaswani
et al., 2017). We computed self-attention using
the torch.bmm() batch multiplication function
implementing the formulae:
Attention(Q, K,V ) = so f tmax(
QK
T
√
n
)V, (3)
where the values for query (Q), key (K), and value
(V) were identical n dimensional data. We also tested
multihead attention with the help of the function
torch.nn.MultiheadAttention. In all cases the number
of heads were 4. To investigate their effects we tested
different variants:
• Self-attention (SA) at the end of the streams;
• Multihead attention (MHA) at the end of the
streams;
• Multihead attention after the concatenation
(fusion) of streams (FMHA).
Figure 6 illustrates these variants.
Figure 6: In our experiments we used different setups for
placing attention and multihead attentions.
5 EXPERIMENTS
5.1 Experiments on CURE
We adopted the standard 60-20-20 split ratio for
training, validation, and test sets, respectively. All
images were resized to the size of 224 × 224, and
pixel values were normalized to the range between
0 and 1. Only user grade images of CURE were
used during training. Training the streams involved
the following hyper-parameter settings: we employed
the Adam optimizer, set the learning rate for all
four streams to 1 ×10
−4
; we applied weight decay
regularization with a coefficient of 1 ×10
−5
; batch
size was 32; margin for the triplet loss function was
chosen 0.5. We trained each model for a total of 30
epochs, and only the best weight file was saved.
As for the fusion phase, we applied different
hyper-parameters: batch size was increased to 128,
since we trained our network on the hard samples
only; learning rate, for better stability, was changed to
2×10
−4
; weight decay was initialized as 1×10
−8
. In
this phase we implemented learning rate scheduling,
which involved the adjustment of the initial learning
rate at every 5 epochs using a gamma value of 0.1.
The network was trained for 30 epochs. Again only
the best weight file was saved.
For all model variants, the training and testing was
accomplished on an NVidia Quadro RTX 5000 GPU.
We replaced the last linear classifier layer of
the EfficientNet models with a new custom linear
layer to satisfy our requirements for the given task.
The number of parameters, after this modification,
for each model was as follows: EfficientNet-B0
with self-attention contained 4.2M parameters,
while EfficientNetV2 S with self-attention and
EfficientNetV2 S with multihead attention involved
20.3M and 20.4M parameters, correspondingly.
In our experiments we followed the standard
procedure: the query image is running through
the embedding process and then the embedding
vector is compared to the embedding vectors of
reference pills. Results are ranked to get Top-1
and Top-5 accuracy. Segregated tests were made
which means that the testing classes were not
included in the training process (except for the
recognition with YOLO, where it is not possible to
run such segregated classifications contrary to pill
localization). Embedding vectors are compared with
Euclidean distance and Top-1 and Top-5 accuracy
values are computed.
The following configurations were evaluated:
• EffNetV1+SA: EfficientNet-B0 and separated
self-attention.
Pill Metrics Learning with Multihead Attention
137

• EffNetV2+SA: EfficientNetV2 S and separated
self-attention.
• EffNetV2+MHA: EfficientNetV2 S and separated
multihead attention.
• EffNetV2+MHA+FMHA: EfficientNetV2 S,
separated multihead attention and multihead
attention in the fusion network.
• EffNetV2+MHA+FMHA+BA: EfficientNetV2 S,
separated multihead attention, multihead attention
in the fusion network, and batch all (BA) strategy
for the fusion network.
In all cases we trained the stream networks with
randomly selected triplets, and the fusion network
with hard triplets (see Eq. 2). The only exception
is EffNetV2+MHA+FMHA+BA when we used the
same random samples for the fusion network as for
the streams.
Results in Table 2 show continuous improvement
of accuracy as the models were incrementally
modified. The largest improvement came from
replacing EfficientNet B0 with EfficientNet V2 S,
which is somehow expected since the number of
parameters is approx. quadrupled.
Table 2: CURE dataset results - two-sided tests.
Top-1 Top-5
EffNetV1+SA 87.18 94.33
EffNetV2+SA 89.23 96.76
EffNetV2+MHA 89.73 97.01
EffNetV2+MHA+FMHA 89.81 97.08
EffNetV2+MHA+FMHA+BA 89.88 97.12
5.2 A Use-Case on OGYEI Dataset
We split the whole set into 70-15-15 parts for training,
validation, and testing. We ran two kinds of tests:
• The same models were tested as in the previous
subsection. We applied the CURE pre-trained
models directly on the OGYEI test images
(segregated test, without any training on OGYEI).
• We evaluated the default YOLOv7 (large)
model containing 306 layers and almost 37M
parameters. Since YOLO requires large number
of training images for each class, we followed
a standard training, validation, and testing
procedure (non-segregated classes for training
and testing).
As for YOLOv7, we utilized the following settings:
images were down-scaled to the image size of 640
× 640, the number of epochs was chosen as 300,
batch size was set to 64. The default on-the-fly
augmentation process of the YOLO’s data loader was
also applied. The training process took about 21 hours
on a Nvidia Quadro RTX A6000 GPU card with 48
GB VRAM. Inference time threshold levels were left
as default: IoU threshold was 0.45 and confidence
threshold was set 0.25. During the evaluation, YOLO
failed to find the pill only on 1 out of 472 images,
while recognition of pills were 435 in the Top-1 and
443 in the Top-5. The inference time took 4-5 ms per
image.
Regarding our multi-stream networks, while the
accuracy values are higher in this test, we observed
very similar improvements as experienced in the
previous subsection: results in Table 3 show
continuous improvement of accuracy as the models
were incrementally modified.
Table 3: OGYEI dataset results - two-sided tests.
Top-1 Top-5
YOLOv7 92.16 93.85
EffNetV1+SA 95.34 99.57
EffNetV2+SA 96.18 100.0
EffNetV2+MHA 96.19 100.0
EffNetV2+MHA+FMHA 96.24 100.0
EffNetV2+MHA+FMHA+BA 96.31 100.0
6 CONCLUSIONS
We followed the strategy of a previous winner
approach for pill recognition in the framework of pill
metrics learning. We introduced different changes
to the original model and evaluated the performance
of the different modifications. We ran all the tests
starting with the full size images and used a trained
model, created on segregated classes, for localization.
The highest Top-1 accuracy could reach 89.88 in
two-sided tests on CURE. It is difficult to compare
our results to others’ since they use different datasets,
different split of the datasets, or even neglect the
localization steps of the pills on input images. While
our split of the CURE dataset (to train, validate, and
test) is slightly different than in (Ling et al., 2020), our
results are consistently larger than the 84.6% Top-1
accuracy given in the supplementary material of (Ling
et al., 2020).
Investigating the misclassified images we found
that some are really hard to recognize (see the first
line of Figure 7) while others should have been solved
(the second line of the same figure). In the future,
we plan to further develop our model with modified
triplet mining and modified triplet loss, as well as the
use of different lighting options.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
138

Figure 7: First line: A query image and the corresponding
wrongly recognized item from the OGYEI dataset. The pill
on the right has similar, but not identical, printed text on
the invisible side. Second line: The same kind of examples
from the CURE dataset.
ACKNOWLEDGEMENTS
This work has been partly supported by the
2020-1.1.2-PIACI-KFI-2021-00296 project of the
National Research, Development and Innovation
Fund. We also acknowledge the financial support
of the Hungarian Scientific Research Fund grant
OTKA K-135729. We are grateful to the NVIDIA
corporation for supporting our research with GPUs
obtained by the NVIDIA Hardware Grant Program.
REFERENCES
Busta, M., Neumann, L., and Matas, J. (2017).
Deep textspotter: An end-to-end trainable scene
text localization and recognition framework. In
Proceedings of the IEEE International Conference on
Computer Vision, pages 2204–2212.
Cimpoi, M., Maji, S., Kokkinos, I., Mohamed, S., , and
Vedaldi, A. (2014). Describing textures in the wild.
In Proceedings of the IEEE Conf. on Computer Vision
and Pattern Recognition (CVPR).
Cronenwett, L. R., Bootman, J. L., Wolcott, J., Aspden, P.,
et al. (2007). Preventing medication errors. National
Academies Press.
Hassan, T. and Khan, H. A. (2015). Handwritten bangla
numeral recognition using local binary pattern.
In 2015 International Conference on Electrical
Engineering and Information Communication
Technology (ICEEICT), pages 1–4. IEEE.
Heo, J., Kang, Y., Lee, S., Jeong, D.-H., and Kim, K.-M.
(2023). An accurate deep learning–based system for
automatic pill identification: Model development and
validation. J. Med. Internet Res., 25:e41043.
Ling, S., Pastor, A., Li, J., Che, Z., Wang, J., Kim, J.,
and Callet, P. L. (2020). Few-shot pill recognition.
In Proceedings of the IEEE/CVF Conference on
Computer Vision and Pattern Recognition, pages
9789–9798.
Liu, L., Zhang, H., Feng, A., Wan, X., and Guo, J.
(2010). Simplified local binary pattern descriptor
for character recognition of vehicle license plate. In
2010 Seventh International Conference on Computer
Graphics, Imaging and Visualization, pages 157–161.
IEEE.
Nguyen, A. D., Nguyen, T. D., Pham, H. H., Nguyen, T. H.,
and Nguyen, P. L. (2022). Image-based contextual pill
recognition with medical knowledge graph assistance.
In Asian Conference on Intelligent Information and
Database Systems, pages 354–369. Springer.
Ojala, T., Pietikainen, M., and Harwood, D. (1994).
Performance evaluation of texture measures with
classification based on Kullback discrimination of
distributions. In Proceedings of 12th International
Conference on Pattern Recognition, volume 1, pages
582–585. IEEE.
Ronneberger, O., Fischer, P., and Brox, T. (2015).
U-Net: Convolutional networks for biomedical image
segmentation. In Medical Image Computing and
Computer-Assisted Intervention–MICCAI 2015: 18th
International Conference, Munich, Germany, October
5-9, 2015, Proceedings, Part III 18, pages 234–241.
Springer.
Schroff, F., Kalenichenko, D., and Philbin, J. (2015).
Facenet: A unified embedding for face recognition
and clustering. In Proceedings of the IEEE
Conference on Computer Vision and Pattern
Recognition, pages 815–823.
Tan, L., Huangfu, T., Wu, L., and Chen, W. (2021).
Comparison of RetinaNet, SSD, and YOLOv3 for
real-time pill identification. BMC Medical Informatics
and Decision Making, 21:1–11.
Tan, M. and Le, Q. (2019). EfficientNet: Rethinking
model scaling for convolutional neural networks.
In International Conference on Machine Learning,
pages 6105–6114. PMLR.
Tan, M. and Le, Q. (2021). EfficientNetv2: Smaller models
and faster training. In International Conference on
Machine Learning, pages 10096–10106. PMLR.
VAIPE (2008). VAIPE-Pill: A Large-scale, Annotated
Benchmark Dataset for Visual Pill Identification.
https://vaipe.org/. [Online; accessed 1-July-2023].
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in Neural
Information Processing Systems, 30.
Wang, C.-Y., Bochkovskiy, A., and Liao, H.-Y. M.
(2023). YOLOv7: Trainable bag-of-freebies sets
Pill Metrics Learning with Multihead Attention
139

new state-of-the-art for real-time object detectors.
In Proceedings of the IEEE/CVF Conference on
Computer Vision and Pattern Recognition, pages
7464–7475.
Yaniv, Z., Faruque, J., Howe, S., Dunn, K., Sharlip, D.,
Bond, A., Perillan, P., Bodenreider, O., Ackerman,
M. J., and Yoo, T. S. (2016). The National Library
of Medicine pill image recognition challenge: An
initial report. In 2016 IEEE Applied Imagery Pattern
Recognition Workshop (AIPR), pages 1–9. IEEE.
Zeng, X., Cao, K., and Zhang, M. (2017). Mobiledeeppill:
A small-footprint mobile deep learning system
for recognizing unconstrained pill images. In
Proceedings of the 15th Annual International
Conference on Mobile Systems, Applications, and
Services, pages 56–67.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
140
