
Addressing Entity Change in Procedural Ontologies
Tyler Johnson
1
, Mohammed Alliheedi
2 a
, Yetian Wang
3 b
and Robert E. Mercer
1 c
1
Department of Computer Science, The University of Western Ontario, London, Canada
2
Department of Computer Science, Al-Baha University, Al Bahah 65527, Saudi Arabia
3
David R. Cheriton School of Computer Science, University of Waterloo, Waterloo, Canada
Keywords:
Experimental Procedure, Procedural Steps, Biochemical Ontology, Time series, Forward-Backward Sequenc-
ing.
Abstract:
Ontologies model a domain by representing the entities, concepts, and the relations between them. The domain
of interest in this position paper is the biochemistry experimental procedure. These procedures are composed
of procedure steps. These steps represent actions. Actions cause change, a concept being implicitly modelled
in this type of ontology. We argue that entities undergoing change need to be properly captured in the ontology.
The biochemistry procedure Alkaline Agarose Gel Electrophoresis is used to demonstrate the generality of this
procedural ontology.
1 INTRODUCTION
The representational adequacy of an ontology, which
provides entities (also known as individuals), con-
cepts, and relations among them, is determined by its
ability to model any situation that can occur in the
domain being represented. Our focus is the biochem-
istry experimental procedure domain.
Descriptions of biochemistry experimental proce-
dures exist in scientific writing. These protocols,
which typically involve several steps, are described
in detail in manuals of standard biochemistry exper-
iment procedures (Boyer, 2012; Sambrook and Rus-
sell, 2001).
This position paper builds upon previous work by
(Alliheedi et al., 2020) which describes ontologies for
two experimental procedures that exist in the man-
ual of standard biochemistry experimental procedures
(Sambrook and Russell, 2001): Alkaline Agarose Gel
Electrophoresis and Southern Blotting. In this po-
sition paper, we provide evidence that to design an
ontology that is adequate for this domain, proper la-
belling of entities that change is required. In addition,
we make explicit the connection to a measurement
ontology and indicate that the modifications conform
to two other ontologies: the geospatial ontology and
a
https://orcid.org/0000-0002-8434-3048
b
https://orcid.org/0000-0002-6984-7256
c
https://orcid.org/0000-0002-0080-715X
BFO. To demonstrate these modifications, we show a
portion of one example of a biochemistry experimen-
tal procedure, Alkaline Agarose Gel Electrophoresis.
2 RELATED WORK
The OBI ontology has been the foundation for several
ontologies in the field of biochemistry procedures,
including those proposed by (Courtot et al., 2008;
Brinkman et al., 2010; Zheng et al., 2013; Soldatova
et al., 2013; Dumontier et al., 2014). These ontologies
are of great interest to our research, and we provide a
brief description of them in this section.
Soldatova and King (Soldatova and King, 2006)
proposed EXPO, an ontology of scientific experi-
ments, which offers a detailed description of vari-
ous aspects of scientific experiments and their re-
lationships. In contrast, OBI describes experimen-
tal processes and relationships, and Brinkman et al.
(Brinkman et al., 2010) discuss three real-world ap-
plications that provide relevant input/output relations
for our purpose. Similarly, Zheng et al. (Zheng et al.,
2013) introduced the beta cell genomics application
ontology (BCGO), which also uses OBI to describe
experimental processes but is primarily a descriptive
ontology. However, some of the relations in RO, the
relation ontology (Smith et al., 2005), that are used in
BCGO (e.g., produces, translate to) have an ordering
sense that aligns with our research.
280
Johnson, T., Alliheedi, M., Wang, Y. and Mercer, R.
Addressing Entity Change in Procedural Ontologies.
DOI: 10.5220/0012239900003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 2: KEOD, pages 280-287
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

Exact (Soldatova et al., 2013) and the Semantic-
science Integrated Ontology (Dumontier et al., 2014)
are the two ontologies that are most similar to our
work. While both aim to describe sets of actions in
scientific protocols, (Alliheedi et al., 2020) represent
sequences of actions. Therefore, relations that de-
scribe orderings of actions (e.g., ‘precedes’ (Dumon-
tier et al., 2014)) are not applicable to sequences since
these relations are transitive. In addition to these on-
tologies, the Molecular Methods Database (MolMeth)
(Klingstr
¨
om et al., 2013) contains scientific protocol
ontologies that conform to a set of laboratory proto-
col standards. Other ontologies that are useful for a
biochemistry procedure-oriented ontology include the
ontologies for processes, such as (Lenat et al., 1985;
Schlenoff et al., 2000), ontology for units of measure
(Rijgersberg et al., 2013), classification of scenar-
ios and plans (CLASP) (Devanbu and Litman, 1996),
and materials ontology (Ashino, 2010). Foundational
theories, such as process calculus and regular gram-
mar, are essential for the formalization of procedure-
oriented ontologies. The current paper builds upon
previous work by (Alliheedi et al., 2020) and provides
a detailed representation of change and an explicit
connection to the measurement ontology (Rijgersberg
et al., 2013).
3 PROCEDURE-ORIENTED
ONTOLOGY
We have proposed in (Alliheedi et al., 2020) a frame-
work for procedure-oriented ontologies. This frame-
work explicitly identifies all steps of an experimen-
tal procedure and provides a set of relations to de-
scribe the relationships between these steps. This
novel approach allows creating a sequence of events
(or steps in a procedure) using the ontological concept
of “something occurs before”. An ontological con-
cept of “sequence” is used to accomplish this. This
new concept is used to model events that happen step
by step in some sort of ordering.
This approach will be used to provide ontologies
representing experimental procedures for Knowledge
Base systems with the required knowledge about the
experimental processes used in biochemistry experi-
mental settings. There are manuals of standard pro-
cedures in biochemistry (Boyer, 2012; Sambrook and
Russell, 2001) which are being used to build exten-
sions of our ontology.
In the next sections, we summarize the current on-
tological framework found in (Alliheedi et al., 2020).
This enables us to connect the focus of this current
position paper, the representation of entity change, to
this framework.
3.1 Relations
In this section, we describe various properties to sat-
isfy the definition of an experimental procedure. An
experimental procedure consists of a series of events,
that is, steps in the procedure. These steps occur in
either partial or total order. Partial ordering allows
steps (more than one step) to precede or follow an-
other step. Total ordering means that a step precedes
or follows another step, and this relation is intransi-
tive. These relations are defined for OWL (McGuin-
ness and van Harmelen, 2004)
1
We represent the choices of a subsequence of steps
from more than one possible subsequence. Since the
choices among subsequences would be “either” or
“or”, the relation ‘optionalStepOf’ needs to be de-
signed based on the different choices of available sub-
sequences in that particular step. To illustrate, the
‘optionalStepOf’ relation is simply an ‘exclusive or’
if there are two choices available, or else it would
be a generalization of the exclusive or. We have im-
plemented the aforementioned relations to satisfy the
definition of “procedure”.
3.2 Classes and Properties
The ontology framework described in detail in (Alli-
heedi et al., 2020) consists of three core classes: Step,
State, and Action. These classes are described in the
following sections. Class names are indicated with
capitalized words, e.g., Step. Properties are indicated
with single quotes, e.g., ‘subStepOf’. Instance names
use typewriter font, e.g., step1. When referring to
the actual steps from the protocol, normal font is used,
e.g., step 1.1.
3.2.1 Step
Each step in a procedure is represented by instances
of the Step class (see Figure 1). We defined object
properties such as ‘precedes’, ‘follows’, and ‘paral-
lel’ to represent the ordering relations of each step.
Note that the aforementioned object properties are
transitive. The properties ‘precedes’ and ‘follows’,
inverses of each other, indicate the chronological or-
der between two steps. The property ‘parallel’ is sym-
metrical which indicates steps may occur simultane-
ously. Figure 1 illustrates a scenario with parallel
1
Available at http://www.ontologydesignpatterns.org/
cp/owl/sequence.owl
Addressing Entity Change in Procedural Ontologies
281
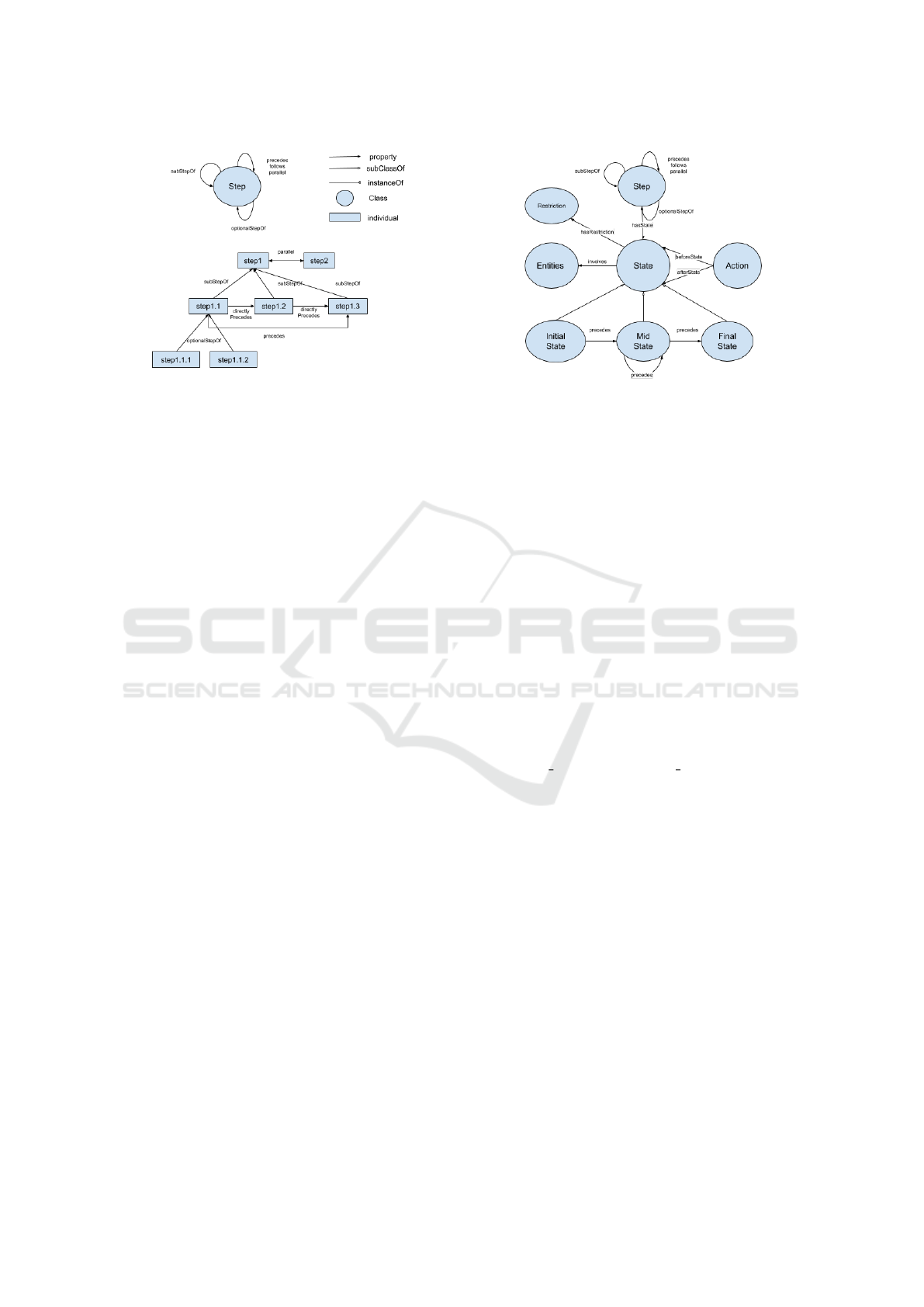
Figure 1: Step class and example instances. (This figure is
from (Alliheedi et al., 2020)).
steps step1 and step2. Intransitive properties ‘di-
rectlyPrecedes’ and ‘directlyFollows’ are subproper-
ties of ‘precedes’ and ‘follows’ respectively. These
properties describe the order between steps in which
a step immediately precedes or follows another step.
Similar to ‘precedes’ and ‘follows’, they are also in-
verses of each other. Therefore, by stating step1.1
‘directlyPrecedes’ step1.2, and step1.2 ‘direct-
lyPrecedes’ step1.3, a reasoner will automatically
infer that step1.1 ‘precedes’ step1.2 as well as
step1.3. Also, step1.3 ‘directlyFollows’ step1.2
but only ‘follows’ step1.1, both being inferable by
a reasoner.
Procedures are often formed by an hierarchical
structure among steps, that is, a step may consist of a
number of sub-steps required to complete. The prop-
erty ‘subStepOf’ indicates that some step(s) must be
completed for the completion of the parent step. Fig-
ure 1 shows an example of a step called step1 which
consists of step1.1, step1.2, and step1.3. Then
step1.1, step1.2, and step1.3 must be completed
in order to state that step1 is complete. The sub-step
step1.1 must complete before step1.2 and thus be-
fore step1.3. The figure also shows the property
‘optionalStepOf’. In the case where one and only one
of step1.1.1 and step1.1.2 needs to be completed
in order to complete step1.1, the property ‘option-
alStepOf’ can be used to indicate that one of the steps
(not both) must be completed in order to complete the
parent step. Both domain and range of the properties
are the class Step.
3.2.2 State
The relations between instances of the class Step out-
line the structure of a procedure. Each instance of
Step is represented as a set of states and are associ-
ated to a set of actions. A step involves a transition
from state to state via a single or a series of actions,
Figure 2: State and Action classes. (This figure is from
(Alliheedi et al., 2020)).
represented by the classes State and Action (see Fig-
ure 2).
The State class is connected to the Step class via
the property ‘hasState’. It has three subclasses, Ini-
tialState, MidState, and FinalState. The subclasses
are connected via properties that include ‘precedes’
and ‘follows’. InitialState can only precede a Mid-
State or a FinalState. FinalState can only follow an
InitialState or a MidState. MidState can ‘follow’
an InitialState and ‘precede’ a FinalState, as well as
‘precede’ or ‘follow’ another MidState. The triple
(stateX, ‘precedes’, stateY) implies (stateY, ‘fol-
lows’, stateX), ‘follows’ being an inverse property
of ‘precedes’. Figures 1 and 2 omit ‘follows’ to keep
the figures clean. Note that a step has at most one in-
stance of InitialState or FinalState but may have mul-
tiple instances of MidState. For example, an instance
of Step, step1, may involve two instances of State,
i.e., step1 state1 and step1 state2, represented
by the following triples: (step1, hasState, state1),
(step1, ‘hasState’, state2), (state1, ‘precedes’,
state2), in which state1 and state2 are instances
of InitialState and FinalState respectively. The State
class also connects to classes Restriction and Enti-
ties, which are discussed in Section 3.2.4 and 3.2.5,
respectively.
3.2.3 Action
States are connected to the Action class via
‘beforeState’ and ‘afterState’, representing the states
before and after an action, respectively. In other
words, an instance of Action would transition an in-
stance of State to another. For example, when an ac-
tion action1 is performed in state1, state1 will
be modified and thus transitioned into a new state
state2. This would be represented by the triples
(action1, ‘beforeState’, state1) and (action1,
‘afterState’, state2).
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
282

Figure 3: Demonstration of Entities class. This figure is
from (Alliheedi et al., 2020).
3.2.4 Restriction
A Restriction class was created to represent cer-
tain limitations applied to a state in a step. It is
linked to the class State via ‘hasRestriction’. For
example, (state1,‘hasRestriction’,restriction1)
means that restriction1 will be checked against
state1. It may be sufficient to apply a restriction to
a FinalState but it is also possible to apply restrictions
to all states in a step.
3.2.5 Entities and Biochemistry Domain
Knowledge
The State class is connected to the Entities class (see
Figure 3) via the property ‘involves’. Domain knowl-
edge of biochemistry can be described by extending
the Entities class with subclasses such as Instruments,
Materials, and Devices involved in a specific state.
For demonstration purposes, we have included only
selected general concepts related to experimental pro-
cedures described in Section 4. The class Instrument
includes Container and Device where Container ‘con-
tains’ Material which is a class for Chemical and Non-
Chemical materials used in biochemistry experiment
procedures. Compound materials and assembled in-
struments are represented using the property ‘consist-
sOf’. Instrument and Material can be connected to
the class Measure which is a combination of numer-
ical values and Unit of Measure, e.g., 10m is a mea-
sure where the value is 10 with a unit of measure of
meter (Rijgersberg et al., 2013). The Measure class
was extended with subclasses to represent absolute
measures (e.g., 10m), range values (e.g., 5m-10m), and
ratio (e.g., 1/2).
4 ITEM LABELLING
The main contribution of this position paper is to pro-
vide direction in how to represent the changes in en-
tities during the experimental procedure in the ontol-
ogy. We use the Alkaline Agarose Gel Electrophore-
sis procedure (see Figure 4) to demonstrate our ideas.
We have generated a timeline to follow the changes in
all of the entities found in this procedure. A portion of
this timeline is given in Figure 5. We will now detail
the new items in this timeline.
4.1 Details of the Timeline
The timeline in Figure 5 shows our proposed method
for tracking changes to entities in the procedure on-
tology. First, entities have “identifiers” (e.g., item1).
The relation ‘alias’ is used when an entity changes
its identifier. The relation ‘becomes’ is used to in-
dicate a change in some property of an entity. Ex-
amples of these properties are temperature (e.g., the
slurry in the flask is heated) or a change in what
constitutes the entity (e.g., the empty flask is filled
with slurry). We have adopted a naming scheme
for the entities. The name contains the item name,
the step in the procedure, and a short description of
the property change. An example of this naming
scheme is item1 step1.1 mixture which becomes
item1 step1.1 mixtureheated.
For entity A to “become” entity B, the following
must be satisfied:
• A and B must have the same identity (we follow
Sider’s view of identity, that is, an entity main-
tains its identity throughout time (Sider, 2003)).
• A measurable change has occurred with respect to
entity A and thus is transitioned to entity B.
• If entity B is a compound material or assembled
instrument based on entity A the ‘becomes’ rela-
tion also has the effect of a ‘consistsOf’ relation
in the reverse direction.
Another relation in the timeline which helps to
track the changes in the entities is ‘consistsOf’. We
take the definition of this relation from the work sum-
marized in Section 3.2.5: this relation connects the
entity which is a compound material or assembled in-
strument with the components that comprise it.
It is important to note that the use of these re-
lations can be subjective (a matter of choice) or
objective (given by the steps in the procedure de-
scription). For example, there is an ‘alias’ relation
between glassbottle1 and Container1 and an-
other between flask1 and Container1. These re-
lations are objectively given in the procedure since
Addressing Entity Change in Procedural Ontologies
283

Alkaline Agarose Gel Electrophoresis
1. Prepare the agarose solution
1.1. Adding the appropriate amount of powdered agarose to a measured
quantity of H2O in either:
1.1.1. An Erlenmeyer flask (Container 1)
1.1.1.1. Loosely plug the neck of the Erlenmeyer flask with Kimwipes
1.1.2. OR a glass bottle (Container 1)
1.1.2.1. Make sure that the cap is loose
1.2. Heat the slurry (Item1) in (Container1) for the minimum time required
to allow all of the grains of agarose to dissolve using either:
1.2.1. A boiling-water bath
1.2.1.1. Check that the volume of the solution (Item 1) has not been
decreased by evaporation during boiling in (Container 1):
1.2.1.1.1. if yes: replenish with H2O in (Container 1)
1.2.1.1.2. If no: do not add H2O in (Container 1)
1.2.2. OR a microwave oven
1.2.2.1. Check that the volume of the solution (Item 1) has not been
decreased by evaporation during boiling in (Container 1):
1.2.2.1.1. if yes: replenish with H2O in (Container 1)
1.2.2.1.2. If no: do not add H2O in (Container 1)
1.3. Cool the clear solution (Item 1) to 55 C.
1.3.1. Add 0.1 volume of 10x alkaline agarose gel electrophoresis buffer in
(Container 1)
1.3.2. And immediately pour the gel (Item 1) into mold (Container 2)
1.4. After the gel (Item 1) is completely set
1.4.1. Mount it (Item 1) in the electrophoresis tank (Container 3)
1.4.2. Add freshly made 1x alkaline electrophoresis buffer until the gel
(Item 1) is just covered.
2. Prepare DNA samples
2.1. Collect the DNA samples (Item 2) by standard precipitation with
ethanol2
2.2. Dissolve the damp precipitates of DNA (Item 2) in 10-20 µl of 1x gel
buffer. (Item 3)
2.3. Add 0.2 volume of 6x alkaline gel-loading buffer
2.3.1. It is important to chelate all Mg2+ with EDTA before adjusting the
electrophoresis samples to alkaline conditions
3. Initiate the electrophoresis
3.1. Load the DNA samples dissolved in 6x alkaline gel-loading buffer into
the wells of the gel (Container 3)
3.2. Start the electrophoresis at < 3.5 V/cm when the bromocresol green has
migrated into the gel approx. 0.5-1 cm; Turn off the power supply, and
place a glass plate on top of the gel in (Container 3) and then continue
electrophoresis until the bromocresol green has migrated
approximately two thirds of the length of the gel in (Container 3).
4. Finalize the experiment
4.1. Process the gel according to one of the procedures either Southern
hybridization by:
4.1.1. Transfer the DNA either:
4.1.1.1. Directly (without soaking the gel) from the alkaline agarose gel to a
charged nylon membrane. Please see Southern Blotting: Capillary
Transfer of DNA to Membranes
4.1.1.2. OR after soaking the gel in neutralizing solution for 45 minutes at
room temperature to either:
4.1.1.2.1. An uncharged nitrocellulose as described in Southern Blotting:
Capillary Transfer of DNA to Membranes
4.1.1.2.2. OR nylon membrane as described in Southern Blotting: Capillary
Transfer of DNA to Membranes
4.1.2. Detect the target sequences in the immobilized DNA by hybridization
to an appropriate labeled probe. Please see Southern Hybridization of
Radiolabeled Probes to Nucleic Acids Immobilized on Membranes
4.2. OR Staining
4.2.1. Soak the gel in neutralizing solutions for 45 minutes at room
temperature
4.2.1.1. Stain the neutralized gel with 0.5 µg/ml ethidium bromide in 1x
TAE or with SYBR Gold
4.2.1.1.1. A band of interest can be sliced from the gel and subsequently
eluted by one of the procedures described Recovery of DNA from
Agarose Gels
Figure 4: The steps of Alkaline Agarose Gel Electrophoresis.
once the type of container is chosen, the proce-
dure continues and performs the same steps using
the container labelled as Container1. On occasion
the choice to use a ‘becomes’ relation is subjective.
For example, item1 step1.3 cooled ‘becomes’
item1 step1.3.1 with10xAgarose and ‘consist-
sOf’ 10xAgarose and item1 step1.3 cooled, by
virtue of the ‘becomes’ relation.
4.2 Measurement Ontology
The ontology presented in Section 3 uses the concept
of measurement. Here, we have made this precise by
incorporating the Ontology of Units of Measure and
Related Concepts (OM) (Rijgersberg et al., 2013).
In Figure 5 connections with the Measurement
Ontology are indicated by green relations and are la-
belled with a label prefixed with “OM:”. With these
explicit measurements, the ontology is able to com-
pare measurements of the changing entities. These
comparisons are indicated by red relations. Some of
the measurements are quantitative (e.g., 55
◦
C) and
some are qualitative (e.g., heated).
To reduce the number of lines in Figure 5, we as-
sume that measurements are transitive with respect to
the ‘becomes’ property. To add, properties will be
transitive unless they are overridden by new measure-
ments of the same type; however, in practice the mea-
surements are not transitive and thus need to be con-
nected to each one that they apply to. For example,
if the temperature does not change in an entity, it will
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
284

Figure 5: Proposed Timeline for the Alkaline Agarose Gel Electrophoresis Procedural Ontology.
Addressing Entity Change in Procedural Ontologies
285

need to be linked back up to the previous measure-
ment of the temperature.
At times, the ontology does not specify a specific
quantity for a measure but allows the use of the on-
tology to determine that quantity. For instance, in
Step 1.1 “appropriate amount of powdered agarose”
and “measured quantity of H2O” will give the desired
concentration to be used in the preparation of the gel
which is determined by the size of the protein being
studied (Lee et al., 2012).
4.3 Other Ontologies
There are other ontologies which seek to map out
a series of events. One ontology is from (Kaup-
pinen and Hyv
¨
onen, 2007; Kauppinen et al., 2008)
where they seek to describe changes in an arbitrary
geospatial region. In particular, they create a ontol-
ogy out of different areas and their changes of Fin-
land since the start of the 20th century. Using what
are called “change bridges”, they connect different
regions as they grow and change throughout time.
In contrast to our work which uses object proper-
ties to link changes in our entities, (Kauppinen and
Hyv
¨
onen, 2007; Kauppinen et al., 2008) uses individ-
uals of a certain type of “change bridge” to represent
what kind of change has occurred in the region(s).
The object properties “before” and “after” are con-
nected from the bridge to the regions to give direction
to the flow of time. This works analogously to the
Action class in Section 3.2.3 with each state in time
linked to an instance of Action by ‘beforeState’ and
‘afterState’.
Another ontology which aims to represent series
of events is the Basic Formal Ontology (BFO) (Arp
et al., 2015). Unlike the previously mentioned ontol-
ogy, this is a top-level ontology which aims to sup-
port information retrieval and analysis of other sci-
entific ontologies across a myriad of individual do-
mains. In particular, we look at the classes of Contin-
uant and Occurant in BFO. The former represents the
concept of continuants. These entities will continue
or persist throughout time. In contrast, occurants are
entities which occur or happen. With respect to the
ontology in Section 3, there are continuants such as
a container or an instrument which can be la-
belled with the appropriate subclasses of Continuant.
Whereas, there are occurants such as the instances of
the Action class, which can be labelled with the ap-
propriate subclasses of Occurant.
5 CONCLUSIONS
In this position paper, evidence is provided showing
that to design an ontology that is adequate for the
biochemistry experimental procedure domain, entities
that change must be properly labelled. We also make
explicit the connection to a measurement ontology
(Rijgersberg et al., 2013) and indicate that the modifi-
cations conform to two other ontologies: time (Kaup-
pinen and Hyv
¨
onen, 2007; Kauppinen et al., 2008)
and BFO (Arp et al., 2015). We demonstrate these
modifications in Figure 5 with a portion of one exam-
ple of a timeline of entity changes in a biochemistry
experimental procedure, Alkaline Agarose Gel Elec-
trophoresis in Figure 4, and how the measurement on-
tology is integrated. We have also done a timeline for
another experimental procedure: Southern Blotting.
Going forward, we will incorporate the timeline
into the ontology classes that have been described in
Section 3. The class hierarchies provided in Section
4 indicate that the presented ontology will accept this
modification. Details how to accomplish this will be
developed in the near future.
ACKNOWLEDGEMENTS
We would like to thank all reviewers for their com-
ments, which improved this paper. This research is
funded by an Undergraduate Student Research Intern-
ship (USRI) from the University of Western Ontario
to T. Johnson, and from The Natural Sciences and
Engineering Research Council of Canada (NSERC)
through a Discovery Grant to R. E. Mercer.
REFERENCES
Alliheedi, M., Wang, Y., and Mercer, R. E. (2020). De-
sign of a biochemistry procedure-oriented ontology.
In Knowledge Discovery, Knowledge Engineering and
Knowledge Management. IC3K 2019, volume 1297 of
Communications in Computer and Information Sci-
ence. Springer.
Arp, R., Smith, B., and Spear, A. D., editors (2015). Build-
ing Ontologies with Basic Formal Ontology. The MIT
Press.
Ashino, T. (2010). Materials Ontology: An infrastructure
for exchanging materials information and knowledge.
Data Science Journal, 9:54–61.
Boyer, R. F. (2012). Biochemistry Laboratory: Modern
Theory and Techniques. Prentice Hall.
Brinkman, R. R., Courtot, M., Derom, D., Fostel, J. M.,
He, Y., Lord, P., Malone, J., Parkinson, H., Peters,
B., Rocca-Serra, P., Ruttenberg, A., Sansone, S.-A.,
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
286

Soldatova, L. N., Jr., C. J. S., Turner, J. A., Zheng, J.,
and the OBI consortium (2010). Modeling biomedical
experimental processes with OBI. Journal of Biomed-
ical Semantics, 1 (Suppl 1):S7.
Courtot, M., Bug, W., Gibson, F., Lister, A. L., Malone,
J., Schober, D., Brinkman, R. R., and Ruttenberg, A.
(2008). The owl of biomedical investigations. In Pro-
ceedings of the Fifth OWLED Workshop on OWL: Ex-
periences and Directions, page 12pp.
Devanbu, P. T. and Litman, D. J. (1996). Taxonomic plan
reasoning. Artificial Intelligence, 84(1-2):1–35.
Dumontier, M., Baker, C. J., Baran, J., Callahan, A., Che-
pelev, L., Cruz-Toledo, J., Rio, N. R. D., Duck, G.,
Furlong, L. I., Keath, N., Klassen, D., McCusker,
J. P., Queralt-Rosinach, N., Samwald, M., Villanueva-
Rosales, N., Wilkinson, M. D., and Hoehndorf, R.
(2014). The Semanticscience Integrated Ontology
(SIO) for biomedical research and knowledge discov-
ery. Journal of Biomedical Semantics, 5(1).
Kauppinen, T. and Hyv
¨
onen, E. (2007). Modeling and rea-
soning about changes in ontology time series. In Shar-
man, R., Kishore, R., and Ramesh, R., editors, On-
tologies, volume 14 of Integrated Series in Informa-
tion Systems, pages 319–338. Springer.
Kauppinen, T., Hyv
¨
onen, E., and V
¨
a
¨
at
¨
ainen, J. (2008). Cre-
ating and using geospatial ontology time series in a
semantic cultural heritage portal. In Bechhofer, S.,
Hauswirth, M., Hoffmann, J., and Koubarakis, M., ed-
itors, The Semantic Web: Research and Applications,
volume 5 of ESWC: European Semantic Web Confer-
ence, pages 110–123. Springer.
Klingstr
¨
om, T., Soldatova, L., Stevens, R., Roos, T. E.,
Swertz, M. A., M
¨
uller, K. M., Kala
ˇ
s, M., Lambrix,
P., Taussig, M. J., Litton, J.-E., Landegren, U., and
Bongcam-Rudlof, E. (2013). Workshop on laboratory
protocol standards for the Molecular Methods Data-
base. New Biotechnology, 30(2):109–113.
Lee, P. Y., Costumbrado, J., Hsu, C. Y., and Kim, Y. H.
(2012). Agarose gel electrophoresis for the separation
of dna fragments. Journal of Visualized Experiments,
62.
Lenat, D. B., Prakash, M., and Shepherd, M. (1985). CYC:
Using common sense knowledge to overcome brittle-
ness and knowledge acquisition bottlenecks. AI Mag-
azine, 6(4):65–65.
McGuinness, D. L. and van Harmelen, F. (2004). OWL
web ontology language overview. W3C Rec-
ommendation, World Wide Web Consortium.
http://www.w3.org/TR/2004/REC-owl-features-
20040210/.
Rijgersberg, H., Van Assem, M., and Top, J. (2013). Ontol-
ogy of units of measure and related concepts. Seman-
tic Web, 4(1):3–13.
Sambrook, J. and Russell, D. W. (2001). Molecular
Cloning: A Laboratory Manual. Cold Spring Harbor
Laboratory Press.
Schlenoff, C., Schlenoff, C., Tissot, F., Valois, J., and Lee,
J. (2000). The Process Specification Language (PSL)
Overview and Version 1.0 Specification. NISTIR
6459, National Institute of Standards and Technology.
Sider, T. (2003). Four-Dimensionalism: An Ontology of
Persistence and Time. Clarendon Press.
Smith, B., Ceusters, W., Klagges, B., K
¨
ohler, J., Kumar, A.,
Lomax, J., Mungall, C., Neuhaus, F., Rector, A. L., ,
and Rosse, C. (2005). Relations in biomedical ontolo-
gies. Genome Biology, 6(5):R46.
Soldatova, L., King, R., Basu, P., Haddi, E., and Saunders,
N. (2013). The representation of biomedical proto-
cols. EMBnet.journal, 19(B).
Soldatova, L. N. and King, R. D. (2006). An ontology of
scientific experiments. Journal of the Royal Society
Interface, 3(11).
Zheng, J., Manduchi, E., and Jr, C. J. S. (2013). Develop-
ment of an application ontology for beta cell genomics
based on the ontology for biomedical investigations.
In 4th International Conference on Biomedical Ontol-
ogy, pages 62–67.
Addressing Entity Change in Procedural Ontologies
287
