
Knowledge Graphs Extracted from Medical Appointment
Transcriptions: Results Generating Triples Relying on LLMs
Rafael Roque de Souza
2 a
, Thiago Luna Pinheiro
2 b
,
Julio Cesar Barbour Oliveira
2 c
and Julio Cesar dos Reis
1 d
1
Precision Data, S
˜
ao Paulo, Brazil
2
Institute of Computing, University of Campinas, Campinas, Brazil
Keywords:
Knowledge Graphs, RDF Triple Generation, eHealth, Telemedicine, Clinical Appointments, LLMs.
Abstract:
Knowledge Graphs (KGs) represent computer-interpretable interactions between real-world entities. This can
be valuable for representing medical data semantically. We address the challenge of automatically transform-
ing transcripted medical conversations (clinical dialogues) into RDF triples to structure clinical information.
In this article, we design and develop a software tool that simplifies clinical documentation. Our solution
explores advanced techniques, such as the Fine-tuned GPT-NeoX 20B model, to extract and summarize cru-
cial information from clinical dialogues. We designed the solution’s architecture, supported by technologies
such as Docker and MongoDB, to be durable and scalable. We achieve accurate medical entity detection from
Portuguese-language textual data and identify semantic relationships in interactions between doctors and pa-
tients. By applying advanced Natural Language Processing techniques and Large Language Models (LLMs),
our results improve the accuracy and relevance of RDF triples generated from clinical textual data.
1 INTRODUCTION
In today’s medical landscape, patient records are a
key source of information. They encompass diag-
noses, medical histories, treatments, and other perti-
nent information. They are crucial to ensuring patient
care quality and continuity. Many of these records
are in unstructured formats, such as handwritten notes
or transcripts of dialogues. The inherent complexity
of medical terminology (Kormilitzin et al., 2021) and
the lack of standardization in data formats make the
analysis and interpretation of these records challeng-
ing (Wu et al., 2020; Honnibal and Johnson, 2015).
This avoids the adequate use and analysis of health-
care data in the ecosystem.
Healthcare professionals, such as doctors, nurses,
and specialists, constantly collect and update patient
health information during appointments. Each inter-
action represents a chance to collect vital data, from
symptoms to test results. Assimilating, interpreting,
and synthesizing this information is crucial to deter-
a
https://orcid.org/0000-0003-1492-5816
b
https://orcid.org/0009-0000-5548-0150
c
https://orcid.org/0000-0002-9990-9016
d
https://orcid.org/0000-0002-9545-2098
mining the next steps in treatment (Velupillai et al.,
2018).
In this context, healthcare professionals demand
further help from technologies to facilitate and aug-
ment their experience. Given the growing amount of
information and the need for standardization, profes-
sionals seek digital software tools that help them or-
ganize, structure, and visualize data. The advanced
digitization of medical records presents challenges in
extracting information from unstructured texts. Nat-
ural Processing Language (NLP) has emerged as a
promising solution, but its application in clinical con-
texts still presents limitations, especially when identi-
fying complex constructions (Sarzynska-Wawer et al.,
2021).
Exploring technologies like NLP can revolution-
ize clinical data management. However, the path to
its effective implementation presents open research
challenges. The search for a solution that harmonizes
efficiency, accuracy, and accessibility persists. The
application of NLP in analyzing Electronic Health
Records Electronic Health Record (EHR) in unstruc-
tured text formats opens the door to quantifying out-
comes that traditionally require detailed abstraction of
the records. To this end, Knowledge Graphs Knowl-
edge Graphs (KGs) can be valuable to structure data
Roque de Souza, R., Pinheiro, T., Oliveira, J. and Reis, J.
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs.
DOI: 10.5220/0012259000003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 2: KEOD, pages 129-139
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)
129

semantically (Kamdar and Dumontier, 2015; Kanza
and Frey, 2019; Ruan et al., 2019). A KGs sys-
tematizes data resources and their interrelationships.
The Resource Description Framework (RDF) serves
as a standard for describing this semantically enriched
data (Candan et al., 2001; Rossanez and dos Reis,
2019).
In this article, we address how structuring clinical
data by combining NLP with KG to transform clin-
ical dialogues into semantic representation via RDF
triples. Our proposed methodology evaluates the
clinical relevance of input textual data (transcripted
from audio records in telemedicine). We explore
advanced techniques for clinical data extraction and
summarization, such as the Fine-tuned Generative
Pre-trained Transformer (GPT)-NeoX 20B model. At
the heart of this innovation, we originally designed
and developed a software solution that simplifies clin-
ical documentation and enhances medical decision-
making.
Our experimental evaluation assesses automatic
clinical text classification in identifying relevant clin-
ical texts from the overall transcriptions. Our solu-
tion explored LLMs and few-show learning for this
purpose. In addition, we present our results of RDF
triple extraction from textual data (relevant clinical
texts). We found relevant findings exploring few-shot
prompting for identifying RDF triples.
The remainder of this article is organized as fol-
lows: Section 2 introduces underlying concepts and
presents the related work. Section 3 details our de-
signed methodology. Section presents key aspects of
our original developed software tool for clinical data
documentation. Section 5 describes our experimen-
tal evaluation and presents the achieved results, which
are discussed in Section 6. Section 7 summarizes our
findings and points out directions for future research.
2 BACKGROUND
KG structured human knowledge modeling the rela-
tionships between real-world entities (Ehrlinger and
W
¨
oß, 2016). They use the RDF triple representation
for KG model. Triples, made up of subject, predicate,
and object, constitute the fundamental structure of
KG. This formal computational representation is es-
sential for describing and understanding information
about diseases. In the context of clinical data, proper
data representation and integration is crucial. It al-
lows healthcare practitioners and researchers to visu-
alize interrelationships between concepts and findings
(Auer et al., 2007), correlating their research with oth-
ers. By observing these relationships, new hypothe-
ses can be formulated, advancing domain knowledge
(Rossanez et al., 2020).
In the biomedical field, KG have been gaining
prominence. Recent initiatives propose innovative
approaches for classification and search strategies,
from user interaction to machine learning. For exam-
ple, studies have converted neuroscience information
into RDF format (Lam et al., 2007), whereas others
have developed frameworks that integrate information
from multiple domains (Rossanez et al., 2020).
In medicine, clinical transcriptions play a crucial
role in documenting clinical information. The digi-
tal revolution has intensified this relevance, convert-
ing clinical dialogues into structured data. This trans-
formation enhances evidence-based decisions and im-
proves the continuity of patient care. However, the
medical language, full of jargon and specific ter-
minology, poses challenges to the analysis of these
transcripts (Exner and Nugues, 2012). Large Lan-
guage Models (LLM) have emerged as a response to
these challenges, improving natural language analy-
sis. LLM have been essential in creating KG in the
biomedical field, combining efficiency and precision
(Lam et al., 2007; Exner and Nugues, 2012; Manning
et al., 2014).
The extraction of RDF triples from texts has be-
come a central issue. In existing studies, techniques
such as Semantic Role Labeling (SRL) are applied to
map entities and determine (Exner and Nugues, 2012)
relationships. However, creating KG from scientific
literature poses challenges. The literature has a par-
ticular and diverse writing style characterized by long
sentences, abbreviations, and technical terms. NLP
tools must be adequately trained for this specific lex-
icon. In this sense, the automatic generation of KG
from scientific literature proves challenging.
Building a KG for all diseases is challenging. For
this reason, DEKGB (Sheng et al., 2019) proposed
an efficient and extensible framework to build KG for
specific diseases based on doctors’ knowledge. They
described the process by extending an existing health
KG to include a new disease.
In this work, we originally explore the potential of
LLMs in generating RDF triples from medical con-
sultation transcripts. We present results that highlight
the effectiveness of our approach and establish a novel
findings for the construction of KG in the medical do-
main.
3 METHODOLOGY
This section describes the conceptual methodology,
as illustrated in Figure 1. We detail the conducted
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
130

research into developing our AIRDoc system. This
research comprised several stages, from data collec-
tion and processing to implementing advanced NLP
techniques. We employed state-of-the-art computa-
tional models like Bidirectional Encoder Represen-
tations from Transformers (BERT) and Fine-tuned
GPT-NeoX to refine our analysis.
In this research, we obtained results and insights
that led us to develop the AIRdoc system (cf. Section
4). This aims to transform medical transcriptions into
organized and intuitive KGs. AIRdoc’s main strength
is its ability to improve accuracy in analyzing medi-
cal dialogues. It is positioned as a relevant software
tool for healthcare professionals, academics, and re-
searchers, supporting decision-making and enhancing
excellence in medical care.
The following presents the methodology used to
build KG from medical consultation transcripts.
3.1 Stage 1: Video/Audio Data
Acquisition
We propose an innovative approach to transform med-
ical transcriptions into KG using the representation of
RDF triples by exploring video and audio input in the
solution. Based on this data acquisition, we use ad-
vanced NLP techniques and machine learning mod-
els focused on accurately detecting entities and iden-
tifying semantic relationships in interactions between
doctor and patient. We then represent these relation-
ships in a KG, providing a hierarchical and organized
view of clinical information.
3.2 Stage 2: Speech Recognition
In the digital age, converting speech into text has be-
come essential for various applications, from virtual
assistants to medical transcriptions. In this stage, we
prioritize the implementation of advanced Speech-to-
Text tools. We calibrate these technological tools to
ensure the highest accuracy in audio transcription, es-
pecially in medical contexts where clarity and pre-
cision are crucial. These technologies have made it
possible to efficiently capture verbal interactions and
convert them into textual records, ready for analysis
and storage.
3.3 Stage 3: Clinical Text Classification
Analyzing medical dialogues requires a meticulous
approach due to the complexity and specificity of
medical terminology. Therefore, at this stage, we ex-
plore and experiment with models to help us classify
clinical texts. On this basis, we use the BERT model
(Devlin et al., 2018), one of the most advanced NLP
architectures. We train BERT to identify and classify
entities in dialogues, such as symptoms, medications,
and diagnoses. With this model, we extracted valu-
able information from the clinical conversations, en-
hancing our understanding of medical interactions.
3.4 Stage 4: Data Extraction and
Structuring
In this stage, we focus on extracting relevant infor-
mation from the data collected. The extraction pro-
cess transforms raw transcripts into structured infor-
mation, such as medications, symptoms, diseases, and
summarizations.
3.4.1 NER
In the Named Entity Recognition (NER) technique,
extracting and categorizing specific information in
medical transcriptions is fundamental. This technique
identifies and categorizes entities in texts, such as
names, places, and temporal expressions. In the med-
ical field, NER highlights terms such as medicines,
diseases, and medical procedures(Neumann et al.,
2019).
In the medical context, the importance of NER is
amplified due to the complexity and specificity of the
terminology used. The correct identification of med-
ical terms, considering Portuguese’s linguistic nu-
ances, is crucial to ensure the accuracy and relevance
of the information extracted. To achieve this pur-
pose, we used pre-trained models in the Portuguese
language, refining them with specific medical data.
This provides greater sensitivity to the clinical con-
text. With the help of NER, we extracted crucial infor-
mation from the transcripts, such as symptoms men-
tioned by the patients, prescribed medications, and
medical histories. This information forms the basis
for subsequent analysis and drawing up KG, essential
for a comprehensive understanding of doctor-patient
interactions.
3.4.2 Summarization
The summarization technique aims to condense infor-
mation from long texts, keeping only the most rele-
vant content. In our context, this technique becomes
fundamental due to the intrinsic complexity of clini-
cal texts. The aim is to uncomplicate medical termi-
nology, facilitate the identification and extraction of
relationships between entities, optimize data process-
ing, and, at the same time, preserve the informative
core of the text.
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs
131

Figure 1: Developed Methodology.
The nature of the input provided to our models
significantly can impact the triples’ quality. Our ex-
perimental study proposes two main approaches to
feeding the models: the first directly uses the con-
tent of the MTS-Dialog dataset
1
, which transcribes
dialogues between doctors and patients. The second
processes and summarizes this input, eliminating col-
loquialisms, redundancies, and other typical speech
features, making the text more concise. The latter ap-
proach arose from the idea that summarization could
improve triple extraction from unstructured text sen-
tences.
3.4.3 RDF Triple Extraction
The task of extracting semantic relationships from
texts is complex and requires precision. Before pro-
ceeding with the triple extraction, the texts under-
went meticulous pre-processing to ensure their qual-
ity and uniformity. We use advanced models, such as
mREBEL and GPT-NeoX Fine-tuned, specifically for
this purpose. These models were trained to identify
and extract semantic relationships from natural lan-
guage texts, culminating in generating RDF triples.
These triples offer structured representations of the
information, simplifying data integration, queries,
and analysis.
3.5 Step 5: Clinical Results
The clinical outcome stage is fundamental in analyz-
ing medical transcripts, focusing on evaluating and in-
terpreting generated data in the clinical context. After
extraction and processing, the RDF triples are thor-
oughly analyzed to discern their clinical significance,
1
https://github.com/abachaa/MTS-Dialog
relating identified entities, such as symptoms and di-
agnoses, to established clinical patterns. The accu-
racy and relevance of the information extracted is vi-
tal; to ensure its reliability, the results are validated
by physician experts, who identify and correct possi-
ble inconsistencies.
4 AIRDoc: AN AI-AUGMENTED
SOFTWARE TOOL FOR
CLINICAL DOCUMENTATION
Architetural Aspects. The AIRdoc software tool
was designed based on a methodology that employs
a sophisticated conceptual architecture to improve in-
teraction in telemedicine and the EHR. This archi-
tecture integrates advances in NLP, database man-
agement, KGs, and state-of-the-art language models.
This might satisfy the intricate demands of the medi-
cal domain. Figure 2 presents our proposed architec-
ture. This figure presents the front end, delineated by
dashed blue lines; the back-end modules are enclosed
within dashed red lines.
Frontend. In the context of our research into con-
verting medical transcripts into KGs, AIRdoc’s fron-
tend interface, as depicted in Figure 3, stands out as a
crucial element. In our proposed design, the defined
interfaces act as a visual window for healthcare pro-
fessionals interact with patients. In addition, we de-
vised interfaces for facilitating exploration and inter-
action with the generated KGs. The design strives for
simplicity and minimalist aesthetics, prioritizing clar-
ity in the presentation of data. In the solution for KG
interaction, elements in the graph, such as nodes and
connections, have different colors, making it more in-
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
132
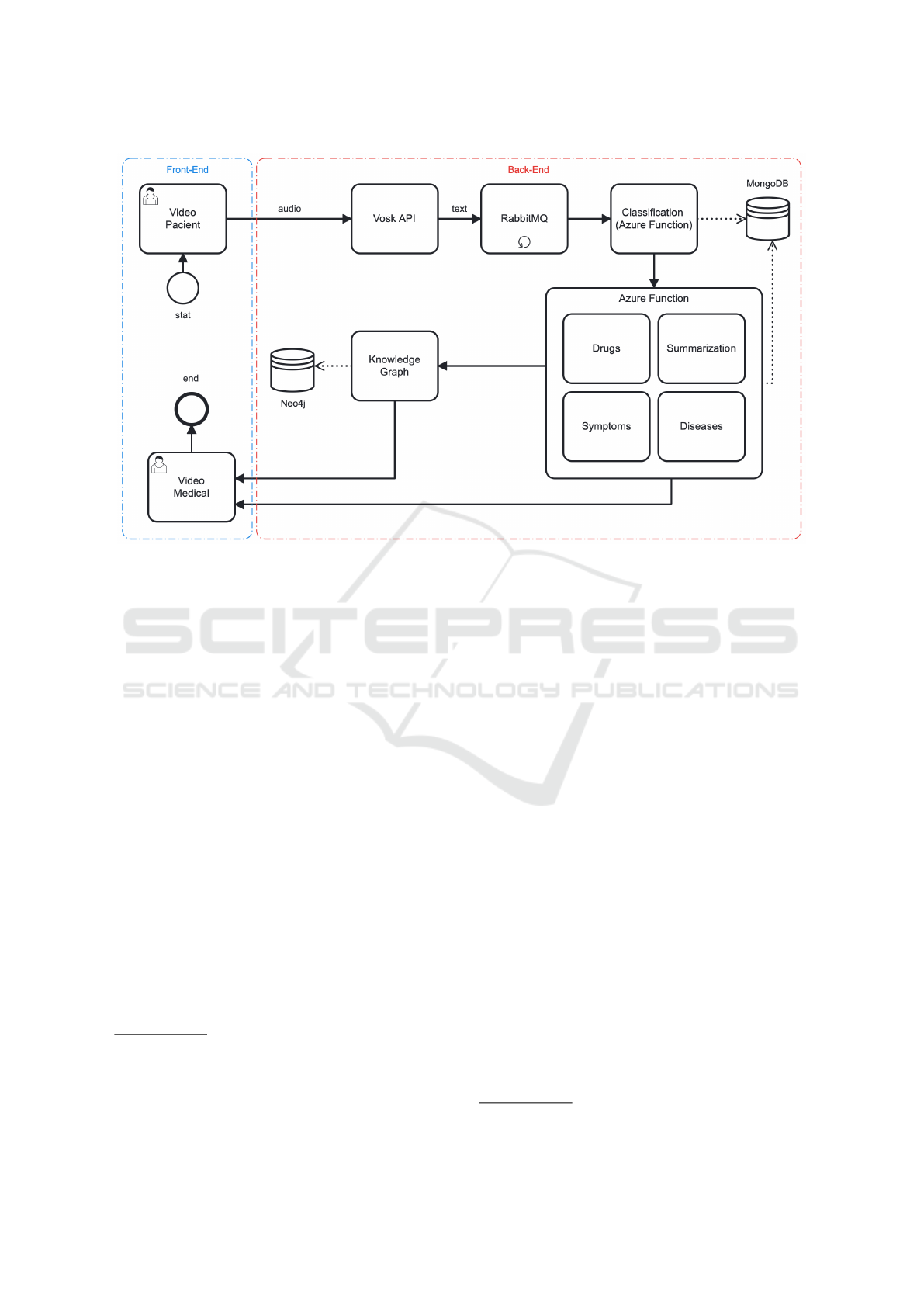
Figure 2: AIRDoc’s Software Architecture for Interactive Construction of Knowledge Graphs.
tuitive to identify and understand the various entities
and their interrelationships from the suggested RDF
triples. This enhances the user experience and aligns
with AIRdoc’s aim of making medical information
more intuitive and easily accessible.
Backend. The conceptual architecture of the
AIRdoc application has been designed, encompass-
ing everything from creating video rooms to advanced
voice capture and real-time transcription techniques.
It is based on database systems such as MongoDB
2
and Neo4j
3
, ensuring efficiency and robustness in
data management. In our implementation, we use
Docker containers, integrated with RabbitMQ
4
, an
advanced messaging system. This shows AIRdoc’s
commitment to providing a cohesive, scalable, high-
performance software environment.
Although AIRdoc’s development began with Java
and Spark, there has been a migration to Python and
FastAPI, reflecting an ongoing drive for greater effi-
ciency and adaptability. This change has enhanced in-
tegration with external platforms, such as NLPCloud
5
and the Vosk
6
API for speech recognition, expanding
its NLP capabilities. Features such as NER, classifi-
cation and summarization of dialogues, and the gen-
eration of RDF semantic triples attests to AIRdoc’s
2
www.mongodb.com
3
https://neo4j.com
4
www.rabbitmq.com
5
https://nlpcloud.com
6
https://alphacephei.com/vosk/
versatility.
Incorporating the KG generator module and the
connection to the Neo4j graph database for storage
purposes underlines AIRdoc’s innovative approach.
These components provide a detailed visual represen-
tation of the interactions between data and ensure a
comprehensive and integrated analysis. In our devel-
opment decisions, we explored the Azure Function
Serverless environment, which offers serverless and
highly scalable execution based on the introduction
of activity logs.
5 EXPERIMENTAL EVALUATION
5.1 Experimental Protocol
Text generation models are machine learning systems
that generate coherent and contextually relevant text
sequences. Recent developments in NLP have estab-
lished these models as essential tools in tasks involv-
ing language understanding and generation.
Model Selection. We began our experimentation
by carefully evaluating available state-of-the-art ma-
chine learning models. After careful analysis, we
chose Fine-tuned GPT-NeoX for assessing the classi-
fication task (accessed via the endpoint of Text Gener-
ation
7
on the NLPCloud platform). For the triple ex-
7
https://docs.nlpcloud.com/#generation
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs
133
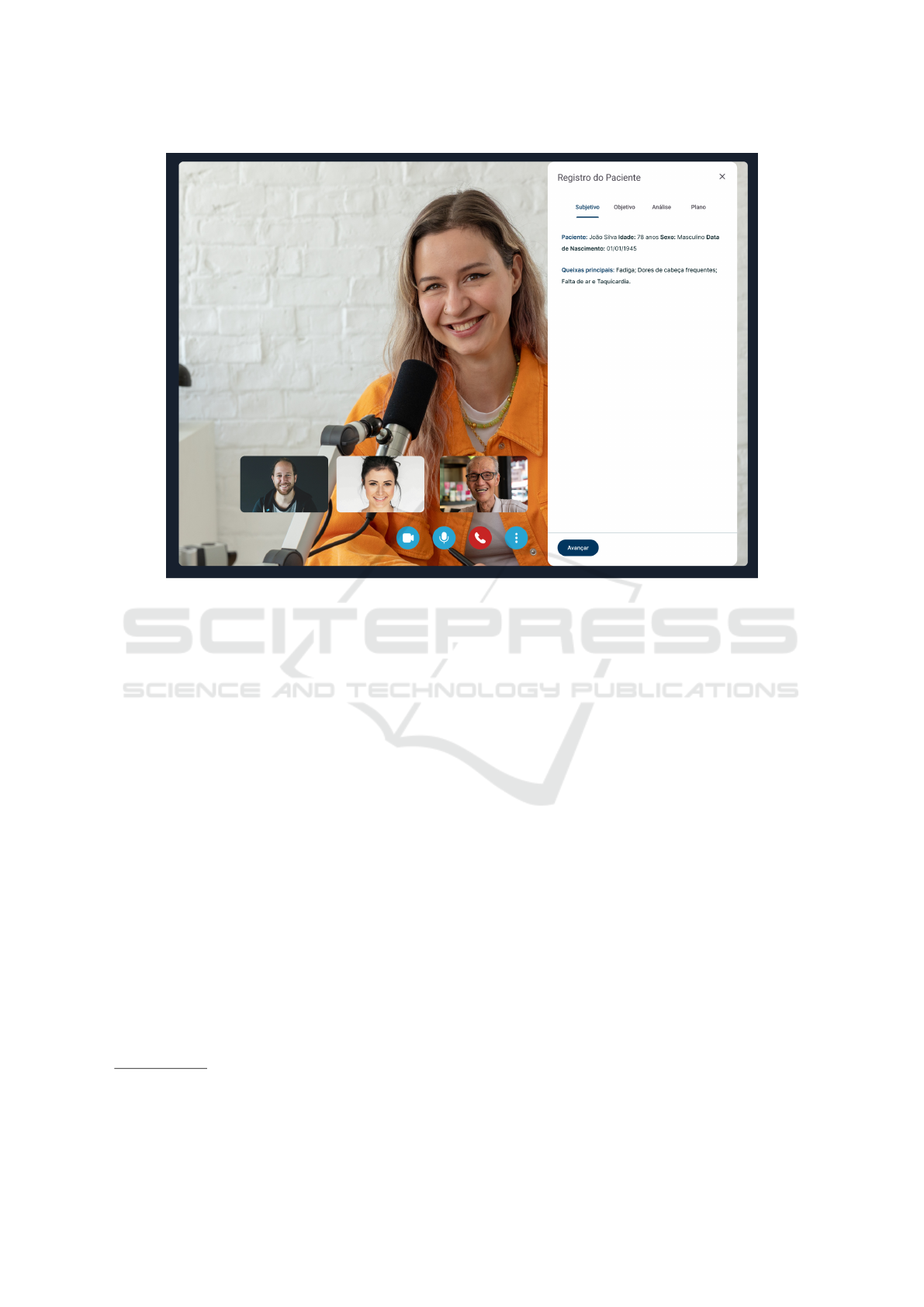
Figure 3: AIRDoc’s Videocall interface for clinical documentation.
traction, we compared the effectiveness of Fine-tuned
GPT-NeoX considering also the mREBEL
8
and the
BLOOM
9
model, bolth from the Hugging Face plat-
form
10
. Our decision was based on the effectiveness,
flexibility, and accessibility of the models.
Definition of Prompts. We clearly defined how to
direct the models for the generation task. This target-
ing, done through a prompt, is essential. We adopted
three strategies to create these prompts:
1. Few-shot prompting. We provided the model
with examples of inputs and their expected out-
puts. In our context, we gave fifteen clinical
sentences and the corresponding triples extracted
from each sentence. These examples are visually
presented in Figure 4. Some cases show the object
acting as the subject, and in others, the predicate
is inferred from the context of the sentence.
2. Simple Instruction. We provided the model
with clear instructions using natural language.
We asked it to extract RDF semantic triples and
present the results in JSON format;
3. Combination of Both. We combined the two pre-
vious strategies, starting with a natural instruc-
8
https://huggingface.co/Babelscape/mrebel-large
9
https://huggingface.co/bigscience/bloom
10
https://huggingface.co
tion followed by examples in the style of few-shot
learning.
Parameters. After defining the prompts, we set
the generation parameters. Two essential parameters
are temperature and top − p. We set temperature to
control the randomness of the output: 0 for Fine-tuned
GPT-NeoX and 0.1 for BLOOM. We aimed to obtain
consistent outputs. For top − p, we varied its values
between 0.5 and 1 for GPT and between 0.1, 0.5, and
0.9 for BLOOM.
Execution and Evaluation. We ran the models
and compared the outcome generated with the ex-
pected results to assess the effectiveness of the models
and the chosen approach.
Interaction. Based on the results, we reviewed
the setup and made adjustments as necessary. We re-
peated the process until we achieved the desired re-
sults.
Metrics. We use four main metrics to evaluate ef-
fectiveness: accuracy, recall, precision and F1-score.
Accuracy gives us an overview of the model’s output
quality. The recall indicates the proportion of posi-
tive (“clinical”) cases correctly classified concerning
the total predicted. Accuracy shows the proportion of
patients correctly classified as positive. The F1-score
represents a harmonic mean between accuracy and re-
call, balancing both metrics.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
134
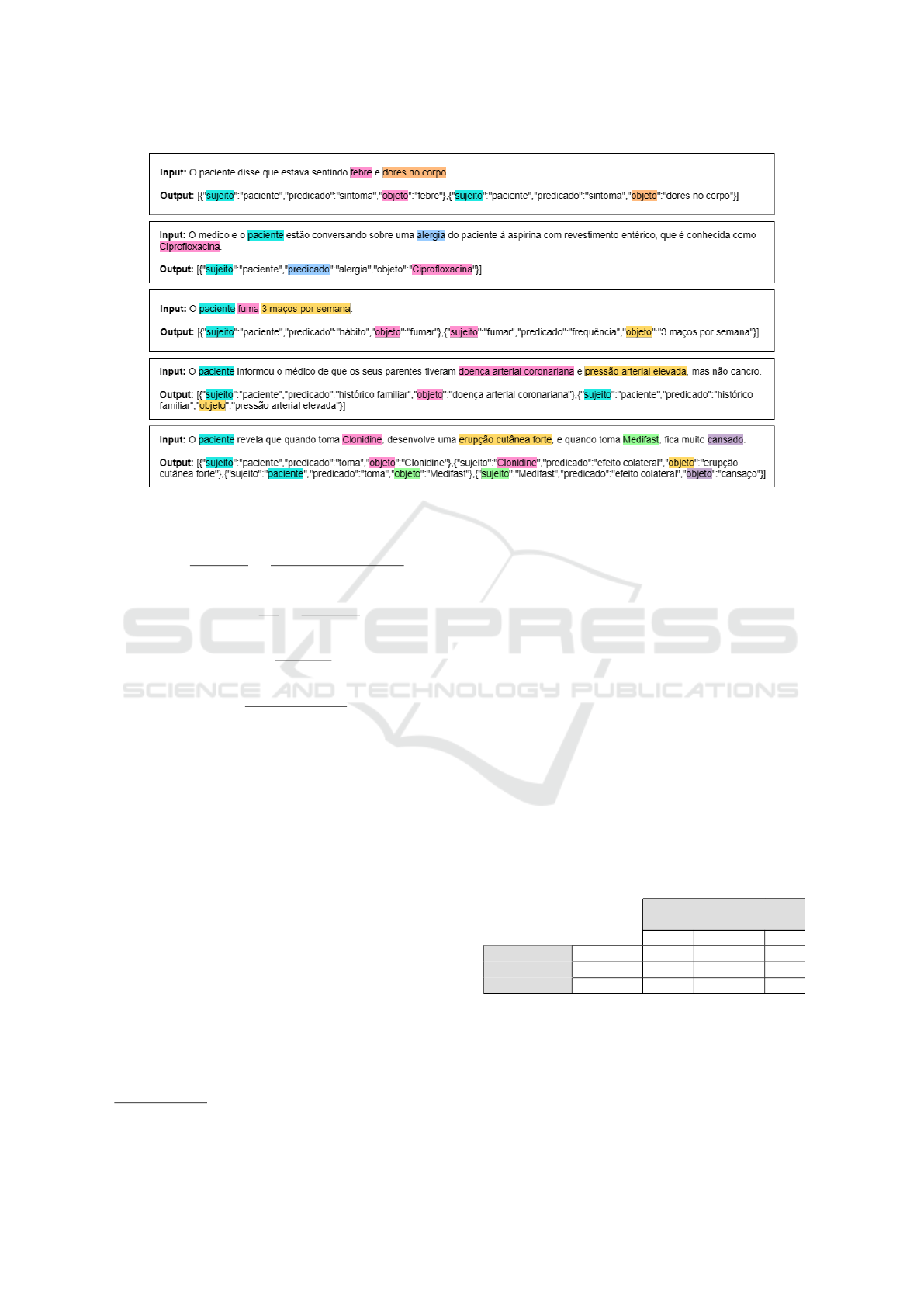
Figure 4: Examples of few-shot prompting where objects can act as subjects or predicates inferred from context.
Ac =
V P + V N
P + N
=
V P + V N
V P + V N + FP + FN
(1)
Recall =
V P
P
=
V P
V P + FN
(2)
Prec =
V P
V P + FP
(3)
F
1
=
2 ∗ Prec ∗ Recall
Prec + Recall
(4)
The equations from 1 to 4 detail the computation
of the metrics. Equation 1 describes Accuracy, fol-
lowed by Recall (Equation 2), Precision (Equation 3)
and F1-score (Equation 4).
5.2 Doctor-Patient Dialogues
We used the MTS-Dialog dataset
11
, translated to
Brazilian Portuguese, as the basis for our experi-
ments. This is composed of dialogues between doc-
tors and patients. This dataset exposed the challenges
associated with the absence of semantic information
in clinical conversations.
5.3 Results of the Clinical Text
Classification
Initially, we analyzed the results from the classifica-
tion stage of 200 doctor-patient dialogues mentioned.
We considered small dialogues with a maximum of
11
https://github.com/abachaa/MTS-Dialog
400 characters. In our evaluation, we assessed both
their original and summarized versions. We evalu-
ated the classification model’s performance by com-
paring its predictions with the classifications in our
previously established Gold Standard dataset.
Table 1 shows the classification of the 200 dia-
logues in their original version, without summariza-
tion, compared to the expected result (Gold Stan-
dard). It classifies the texts into “clinical” and “non-
clinical”, showing the totals for each category. The
analysis shows that the model correctly classified
160 dialogues as “clinical” and 17 as “non-clinical”.
However, a discrepancy was identified in 23 dia-
logues, with 9 cases of false positives and 14 false
negatives. We defined the Positive class for “clinical”
texts and the Negative class for “non-clinical” texts.
Table 1: We compared the gold standard classification with
the one predicted by our model in 200 dialogues between
doctors and patients. We carried out this classification using
the dialogues in their original form.
Prediction
(Original Version)
clinical non-clinical total
clinical 160 14 174
non-clinical 9 17 26
Gold Standard
total 169 31 200
Table 2 shows the results of the evaluation metrics
for classifying the dialogues in their original version.
According to Equation 1, the model correctly classi-
fied 177 of the 200 dialogues evaluated, resulting in
an accuracy of 88.5%. The Recall was 92.5%, ac-
cording to Equation 2. The Precision, determined by
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs
135
Equation 3, was 94.7%. Finally, the F
1
score, calcu-
lated by Equation 4, reached 93.6%.
Table 2: We calculated the evaluation metrics: Accuracy,
Recall, Precision, and F
1
-score for the 200 dialogues be-
tween doctors and patients. We used the dialogues in their
original form to carry out this classification.
Ac Recall Prec F
1
88.5% 92.5% 94.7% 93.6%
Results Based on Summarized Texts. Table 3
presents the analysis focusing on the dialogues af-
ter summarization. The aim is to assess the model’s
ability to distinguish between categories, even with
more summarized information. Of the 200 summa-
rized dialogues, the model categorized 174 as Posi-
tive (P) and 26 as Negative (N). This analysis resulted
in 158 cases of True Positive (VP), 18 of True Nega-
tive (VN), 16 of False Negative (FN), and 8 of False
Positive (FP).
Table 3: We compared the gold standard classification with
the one predicted by our model in the 200 dialogues be-
tween doctors and patients. We used the dialogues in their
summarized form for this classification analysis.
Prediction
(Summarized Version)
clinical non-clinical total
clinical 158 16 174
non-clinical 8 18 26
Gold Standard)
total 166 34 200
Table 4 shows the values of the evaluation met-
rics for classifying the dialogues relying on summa-
rized texts as input for the classification. The model
resulted in an accuracy of 88.0%. The Recall was
91.3%, according to Equation 2. The Precision, de-
termined by Equation 3 was 95.2%. Finally, the F
1
score, calculated by Equation 4, reached 93.2%.
Table 4: We calculated the evaluation metrics: Accuracy,
Recall, Precision and F
1
-score for the 200 dialogues be-
tween doctors and patients. We used the dialogues in their
summarized form for this classification.
Ac Recall Prec F
1
88.0% 91.3% 95.2% 93.2%
5.4 Results of the RDF Triples
Extraction
We present the results of the complete execution of
the pipeline. We used a subset of 20 doctor-patient
dialogues in this experiment, randomly selected from
the 200 dialogues from the previous evaluation. We
start by discussing the results of classifying the clin-
ical aspects in this subset and then make a qualita-
tive analysis of the triples generated in the extrac-
tion phase, highlighting the best effectiveness of each
model, among other aspects.
Table 5 compares the classification predicted by
our model with the expected classification in our Gold
Standard. Table 5 shows the results for both strate-
gies: the one that uses the original dialogue and the
one that uses its summarized version. In the analysis,
we observed 13 and 14 cases of True Positive for the
original and summarized versions, respectively, and
3 and 4 instances of True Negative. As for the pre-
dicted classifications, the original version had 1 case
of False Positive and 3 of False Negative. The sum-
marized version presented 0 and 2 instances of these
errors, respectively.
Based on the results in Table 5, we calculated the
Accuracy, Recall, Precision, and F
1
-score metrics for
both strategies. Table 6 shows the results for the strat-
egy that uses dialogues in their original form. In this
context, the accuracy was 80.0%. The Recall reached
81.3%, while the Precision was 92.9%. The F
1
-score
registered 86.7%.
On the other hand, Table 7 presents the results re-
lying on the summarized version of the dialogs. The
Accuracy and Recall reached 90.0% and 87.5%, re-
spectively. Accuracy reached 100%, and the F
1
-score
was 93.3%.
In the qualitative analysis of the RDF triples gen-
erated (relying on these 20 randomly selected di-
alogs), we observed the key effectiveness of the three
selected models.
• mREBEL: Among the models evaluated for triple
extraction, mREBEL performed worst in extract-
ing RDF triples for original and summarized in-
puts. The triples generated by this model were
often incoherent or did not capture the essential
information. Thus, mREBEL could not produce
RDF triples that adequately reflected the patient’s
clinical condition.
• BLOOM: The BLOOM model, when tested with
the few-shot prompting strategy and variation of
the top − p parameter, showed superior perfor-
mance to mREBEL. This produced quality triples
in several assessements. However, the triples
lacked objectivity and standardization in some sit-
uations, especially when the original dialogue was
used as input. The extracted triples were long-
winded in certain cases, using extensive dialogue
segments instead of keywords or concepts. The
“object” field was often affected by this problem
and was not even generated occasionally.
• Fine-tuned GPT-NeoX 20B. This model of the
GPT family stood out in our evaluation, outper-
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
136

Table 5: Comparing the classification of our gold standard with that classified by our model in 20 randomly selected dialogues
between doctors and patients. We performed this classification using the dialogues in their original and summarized form.
Classification
(Original Version)
Classification
(Summarized Version)
clinical non-clinical total clinical non-clinical total
clinical 13 3 16 14 2 16
non-clinical 1 3 4 0 4 4
Gold
Standard
total 14 6 20 14 6 20
Table 6: Evaluation Metrics: Accuracy, Recall, Precision
and F
1
-score based on the 20 randomly selected dialogues
in their original form.
Ac Recall Prec F
1
80.0% 81.3% 92.9% 86.7%
Table 7: Results for the Evaluation Metrics: Accuracy, Re-
call, Precision and F
1
-score using the summarized 20 ran-
domly selected dialogues.
Ac Recall Prec F
1
90.0% 87.5% 100.0% 93.3%
forming the others regardless of the prompt strat-
egy applied. The prompt based on simple natural
language instructions had remarkable results but
also showed a few inconsistencies, especially in
the output formatting. In contrast, the strategies of
few-shot prompting and the combination of few-
shot prompting with simple instruction produced
consistent, high-quality results.
The triples generated were relevant and crucial to
understanding the patient’s clinical condition. The
model performed similarly for original and summa-
rized inputs. In addition, the best effectiveness was
observed when the top − p parameter was set to 0.5.
Figure 5 shows the triples extracted by each model
with an input example in its original and summarized
version. These examples were extracted relying on
the BLOOM model configured with a temperature
parameter of 0.1 and top − p of 0.9. On the other
hand, the Fine-tuned GPT-NeoX model was adjusted
with the temperature and top − p parameters set to
0.0 and 0.5, respectively. Both models employed the
few-shot prompting strategy for these examples.
With the right approach, we found that text gen-
eration models are highly effective for extracting se-
mantic triples from clinical doctor-patient dialogues
in Portuguese Language. The success depends on the
right combination of model selection, prompt defini-
tion, and parameter tuning.
6 DISCUSSION
In the dynamic digital health scenario, telemedicine is
emerging as a key solution, offering accessible med-
ical care and overcoming geographical obstacles. In
this scenario, technological tools such as AIRdoc are
being developed to enrich the experience of health-
care professionals and patients. However, such in-
novations must undergo careful evaluation to confirm
their effectiveness and relevance.
In this study, we explored the ability of sophis-
ticated NLP techniques to structure clinical data, fo-
cusing on generating RDF triples from clinical dia-
logues. The main motivation for this research was
the demand for a more effective and accessible repre-
sentation of clinical information. Our study not only
defined a new standard for the representation of clin-
ical information but also, indicated a potential im-
pact on healthcare, optimizing patient care and clin-
ical decision-making.
We explored and investigated the use of the
mREBEL, BLOOM and GPT-NeoX models adjusted
to deal with the complexity of semantic relationships
in RDF triple extraction task. A qualitative analy-
sis showed that while mREBEL underperformed, the
adjusted GPT-NeoX was the most promising model
from our findings. BLOOM, despite outperforming
mREBEL, still lacked objectivity.
In our experiments, we observed that the summa-
rization technique proved to be valuable for improv-
ing the quality of the generated RDF triples, espe-
cially for dense clinical texts. The ability to synthe-
size information while maintaining its essence was
decisive for the success of the process. The similarity
of the results between the original and summarized
texts attested to the effectiveness of summarization
for RDF triple extraction.
The employed models proved robust in classifica-
tion, achieving an accuracy of over 88% for both orig-
inal and summarized dialogues. A high recall rate
suggested success in identifying clinical texts. The
transition to the NeoX model highlights the ongoing
commitment to improving the platform’s efficiency.
This choice reinforces the need to continually adapt
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs
137
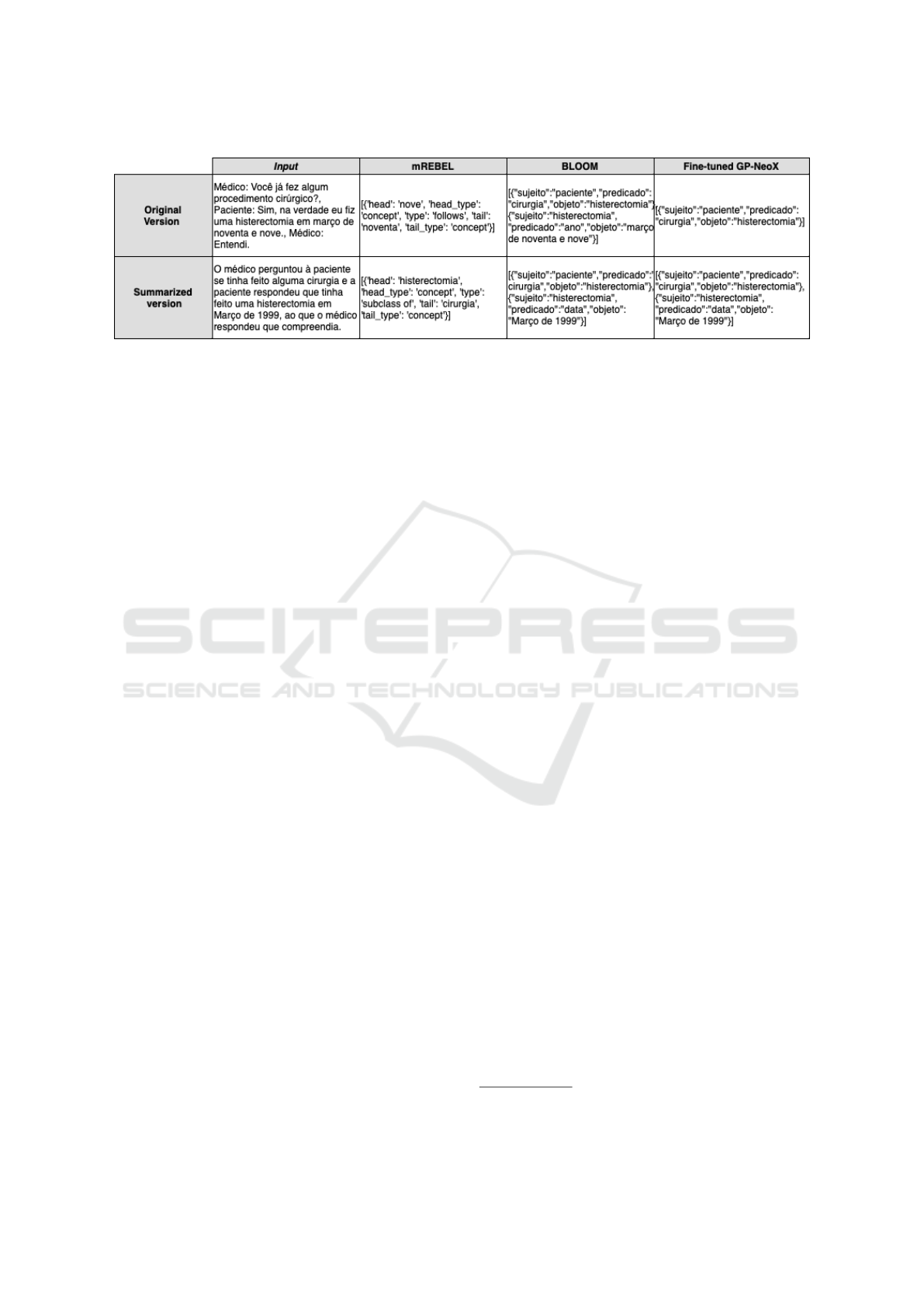
Figure 5: Examples for comparison among the models used for the task of extracting RDF triples (text in Portuguese Lan-
guage).
and evolve the software tool and the underlying tech-
niques adopted. We recognize areas for improvement
in our study. The models’ ability to distinguish be-
tween affirmations, negations, and mentions is chal-
lenging, given the possibility of misinterpretations in
clinical contexts.
In the technological panorama, it is essential to
highlight the role of AIRdoc technology. Even with
its focus on specific NLP models and techniques, the
AIRdoc solution marks a breakthrough in applying
these techniques for clinical data in the Portuguese
language. Integrating advanced NLP models into a
user-friendly interactive solution such as AIRdoc can
advance the current status regarding healthcare pro-
fessionals’ access to and interpretation of clinical in-
formation.
In summary, this study shed light on the poten-
tial and challenges of NLP techniques in structuring
clinical data. The insights gained can reshape the rep-
resentation of clinical information, benefiting profes-
sionals and patients. It is crucial to understand that the
findings are initial and future research is demanded to
further validate and improve the techniques presented.
7 CONCLUSION
This study explored the confluence of advanced NLP
and KG techniques to revolutionize clinical informa-
tion representation, interpretation, and organization.
We developed an innovative method applied to a soft-
ware tool that transforms transcripts from clinical di-
alogues into RDF triples, marking a significant trans-
formation in clinical data representation. We high-
lighted the implementation of a summarization stage,
which proved crucial in highlighting and condensing
essential clinical aspects of the RDF extraction task.
We successfully assessed the effectiveness of the pro-
posed approach for RDF triple generation based on
LLMs and few-shot learning. Results suggested a
promising future for the management and exploration
of semantically structured clinical records. Our found
obstacles reinforce the relevance of future research to
improve the techniques presented. Future studies in-
volve integrating more clinical data sources, improv-
ing summarization accuracy, and expanding the ap-
proach to specific clinical domains.
ACKNOWLEDGEMENTS
This work was supported by the S
˜
ao Paulo Research
Foundation (FAPESP) (Grant #2022/13201-3)
12
.
REFERENCES
Auer, S., Bizer, C., Kobilarov, G., Lehmann, J., Cyganiak,
R., and Ives, Z. (2007). Dbpedia: A nucleus for a web
of open data. In international semantic web confer-
ence, pages 722–735. Springer.
Candan, K. S., Liu, H., and Suvarna, R. (2001). Resource
description framework: metadata and its applications.
Acm Sigkdd Explorations Newsletter, 3(1):6–19.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Ehrlinger, L. and W
¨
oß, W. (2016). Towards a definition
of knowledge graphs. SEMANTiCS (Posters, Demos,
SuCCESS), 48(1-4):2.
Exner, P. and Nugues, P. (2012). Entity extraction: From
unstructured text to dbpedia rdf triples. In WoLE@
ISWC, pages 58–69.
Honnibal, M. and Johnson, M. (2015). An improved non-
monotonic transition system for dependency parsing.
In Proceedings of the conference on empirical meth-
ods in NLP, pages 1373–1378.
Kamdar, M. R. and Dumontier, M. (2015). An ebola virus-
centered knowledge base. Database, 2015:bav049.
12
The opinions expressed in this work do not necessarily
reflect those of the funding agencies.
KEOD 2023 - 15th International Conference on Knowledge Engineering and Ontology Development
138

Kanza, S. and Frey, J. G. (2019). A new wave of innova-
tion in semantic web tools for drug discovery. Expert
Opinion on Drug Discovery, 14(5):433–444.
Kormilitzin, A., Vaci, N., Liu, Q., and Nevado-Holgado,
A. (2021). Med7: A transferable clinical natural lan-
guage processing model for electronic health records.
Artificial Intelligence in Medicine, 118:102086.
Lam, H. Y., Marenco, L., Clark, T., Gao, Y., Kinoshita, J.,
Shepherd, G., Miller, P., Wu, E., Wong, G. T., Liu, N.,
et al. (2007). Alzpharm: integration of neurodegener-
ation data using rdf. BMC bioinformatics, 8:1–12.
Manning, C. D., Surdeanu, M., Bauer, J., Finkel, J. R.,
Bethard, S., and McClosky, D. (2014). The stanford
corenlp natural language processing toolkit. In Pro-
ceedings of 52nd annual meeting of the association
for computational linguistics: system demonstrations,
pages 55–60.
Neumann, M., King, D., Beltagy, I., and Ammar, W.
(2019). Scispacy: fast and robust models for biomed-
ical natural language processing. arXiv preprint
arXiv:1902.07669.
Rossanez, A. and dos Reis, J. C. (2019). Generating knowl-
edge graphs from scientific literature of degenerative
diseases. In SEPDA@ ISWC, pages 12–23.
Rossanez, A., Dos Reis, J. C., Torres, R. d. S., and de Rib-
aupierre, H. (2020). Kgen: a knowledge graph genera-
tor from biomedical scientific literature. BMC medical
informatics and decision making, 20(4):1–24.
Ruan, T., Huang, Y., Liu, X., Xia, Y., and Gao, J. (2019).
Qanalysis: a question-answer driven analytic tool on
knowledge graphs for leveraging electronic medical
records for clinical research. BMC medical informat-
ics and decision making, 19:1–13.
Sarzynska-Wawer, J., Wawer, A., Pawlak, A., Szy-
manowska, J., Stefaniak, I., Jarkiewicz, M., and
Okruszek, L. (2021). Detecting formal thought disor-
der by deep contextualized word representations. Psy-
chiatry Research, 304:114135.
Sheng, M., Shao, Y., Zhang, Y., Li, C., Xing, C., Zhang,
H., Wang, J., and Gao, F. (2019). Dekgb: an extensi-
ble framework for health knowledge graph. In Inter-
national Conference on Smart Health, pages 27–38.
Springer.
Velupillai, S., Suominen, H., Liakata, M., Roberts, A.,
Shah, A. D., Morley, K., Osborn, D., Hayes, J., Stew-
art, R., Downs, J., et al. (2018). Using clinical natu-
ral language processing for health outcomes research:
overview and actionable suggestions for future ad-
vances. Journal of biomedical informatics, 88:11–19.
Wu, S., Roberts, K., Datta, S., Du, J., Ji, Z., Si, Y., Soni,
S., Wang, Q., Wei, Q., Xiang, Y., et al. (2020). Deep
learning in clinical natural language processing: a me-
thodical review. Journal of the American Medical In-
formatics Association, 27(3):457–470.
Knowledge Graphs Extracted from Medical Appointment Transcriptions: Results Generating Triples Relying on LLMs
139
