
DAF: Data Acquisition Framework to Support Information Extraction
from Scientific Publications
Muhammad Asif Suryani
1,2 a
, Steffen Hahne
1 b
, Christian Beth
1 c
, Klaus Wallmann
2 d
and Matthias Renz
1 e
1
Institute of Informatik, Christian-Albrechts-Universit
¨
at zu Kiel, Kiel, Germany
2
GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany
Keywords:
Information Extraction, Data Acquisition, Research Data Management, Scientific Publication, Marine
Science.
Abstract:
Researchers encapsulate their findings in publications, generally available in PDFs, which are designed pri-
marily for platform-independent viewing and printing and do not support editing or automatic data extraction.
These documents are a rich source of information in any domain, but the information in these publications
is presented in text, tables and figures. However, manual extraction of information from these components
would be beyond tedious and necessitates an automatic approach. Therefore, an automatic extraction ap-
proach could provide valuable data to the research community while also helping to manage the increasing
number of publications. Previously, many approaches focused on extracting individual components from sci-
entific publications, i.e. metadata, text or tables, but failed to target these data components collectively. This
paper proposes a Data Acquisition Framework (DAF), the most comprehensive framework to our knowledge.
The DAF extracts enhanced metadata, segmented text, captions and content of tables and figures respectively.
Through rigorous evaluation on two distinct datasets from the Marine Science and Chemical Domain we show-
case the superior performance compared of the DAF to the baseline PDFDataExtractor. We also provide an
illustrative example to underscore DAF’s adaptability in the realm of research data management.
1 INTRODUCTION
Researchers disseminate their findings through scien-
tific publications, most notably in the form of PDF
documents. These PDFs are primarily created for
platform-independent viewing and printing and serve
as repositories of valuable knowledge across diverse
domains (Inc., 2006). However, their primary de-
sign for human readability poses a significant chal-
lenge when it comes to automatically extracting struc-
tured information for further analysis or data manage-
ment tasks. These publications contain valuable in-
sights generally presented in various components, i.e.,
text, tables, and figures. The presented information in
these components is generally densely linked among
the structural components and exhibits diverse char-
a
https://orcid.org/0000-0003-1669-5524
b
https://orcid.org/0009-0008-6434-4001
c
https://orcid.org/0000-0003-3313-0752
d
https://orcid.org/0000-0002-1795-376X
e
https://orcid.org/0000-0002-2024-7700
acteristics. Generally, context elaboration is given
through plain text, while numerical and graphical re-
sults are expressed in tables and figures respectively.
Open access to scientific publications and extract-
ing the relevant data component is a core interest of
research communities in order to learn from previ-
ous findings and acquire new insights from the data
to instigate data-driven research activities (Martinez-
Rodriguez et al., 2020), (Swain and Cole, 2016).
There has been a substantial increase in publica-
tions over the last decades. With the widespread use
of computational methods and the volume and reli-
ability of scientific publications, they can be viewed
as a manifestation of Big Data (Jinha, 2010), (Taylor-
Sakyi, 2016). Hence, the surge in scientific publica-
tions in every domain makes it difficult for researchers
to seek valuable information from these documents
(Zhu and Cole, 2022). Facing the massive volume
of publications, manually extracting information from
them is not a favourable solution, as it is only feasi-
ble for small document corpora. In order to support
research activities in every domain, automatic infor-
468
Suryani, M., Hahne, S., Beth, C., Wallmann, K. and Renz, M.
DAF: Data Acquisition Framework to Support Information Extraction from Scientific Publications.
DOI: 10.5220/0012260300003598
In Proceedings of the 15th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2023) - Volume 1: KDIR, pages 468-476
ISBN: 978-989-758-671-2; ISSN: 2184-3228
Copyright © 2023 by SCITEPRESS – Science and Technology Publications, Lda. Under CC license (CC BY-NC-ND 4.0)

mation extraction approaches are getting increasingly
involved in knowledge discovery and support research
data management activities (Swain and Cole, 2016).
Scientific publications are generally well-written
for human readers but do not support automatic ex-
traction. Besides, an important aspect is that publica-
tions are highly dependent on the templates of their
respective publishers, which is an essential factor to
be considered in the automatic information extraction
process. Hence, to automatically identify and extract
relevant textual, quantitative information, many fac-
tors have to be taken into account, such as layout anal-
ysis, writing styles, variations of expressions or units,
numerical features, and graphical information (Foppi-
ano et al., 2019), (Zhu and Cole, 2022).
Therefore, the automatic extraction of targeted
information from scientific publications is a worthy
idea, which is suitable for researchers to strive for
their desired information by minimizing manual inter-
ventions. The automatic extraction will help in mak-
ing relevant information available digitally. Initially,
numerous studies targeted the extraction of metadata
and bibliographic information from publications, i.e.
DOI, title, authors, journal, abstract, keywords and
references, respectively (Martinez-Rodriguez et al.,
2020). Recently, applications have been introduced,
which focused on the full plain-text from research
publications (Suryani et al., 2022).
Subsequently, in addition to focusing on metadata
and plain-text, it is necessary to incorporate tables and
figures into information extraction spectrum. This in-
clusion will provide access to a wider range of data,
facilitating data-driven research activities.
This paper introduces the Data Acquisition
Framework (DAF), an extensive and comprehensive
solution, which offers a potential solution to manage
the growing volume of scholarly work. DAF focuses
on extracting enriched metadata, segmenting textual
content, and extracting the tables and figures includ-
ing their respective captions. The outcome of DAF’s
rigorous evaluation on two distinct datasets, drawn
from the fields of Marine Geology and Chemistry,
highlights its superiority over the baseline method.
Our contributions can be summarized as follows:
1. We propose a comprehensive Data Acquisition
Framework (DAF) capable of extracting the struc-
tural components, i.e., text, tables and figures,
without template dependency.
2. We propose Document OPTICS, which provides
the document’s structural topology and instigates
an enhancement in traditional metadata.
3. DAF also presents captions for tables and figures.
4. DAF is evaluated on two diverse dataset and
achieve better results than comparable approach.
5. We also showcase a potential information mod-
elling use-case to highlight the efficacy of DAF.
2 RELATED WORK
From scientific publications, extracting structured in-
formation is pivotal for data-driven applications and
knowledge discovery. This section offers a compre-
hensive overview of both individual extraction mod-
ules and entire frameworks, where the focus is on var-
ious facets of data extraction, metadata, textual con-
tent, images, and tables.
2.1 Data Extraction Packages
Researchers have primarily focused on extracting
metadata from scientific publications, e.g. DOIs, au-
thors, titles, and venues. This extracted information is
utilized in various potential applications, such as Bib-
liographic Networks, Recommendation Systems and
Heterogeneous Information Networks (HINs) (Kreutz
and Schenkel, 2022), (Yadav et al., 2019), (Guerra
et al., 2018).
Rcrossref (Chamberlain et al., 2023) and fulltext
(Chamberlain, 2019) are modules in R that extract
metadata from scientific publications online. Both
modules cover numerous publishers and provide,
DOI, title, authors, journals, abstracts, etc. as data-
frame. PDFminer.six (PDFminer, ) is a community-
maintained version of PDFminer, that offers a range
of extraction capabilities from metadata to structural
extraction. Apache Tika (Contributors, b), a Java li-
brary, which is also available as a Python package,
which extracts metadata and plain-text from publica-
tions. The output from Tika covers a relatively high
number of structural features besides metadata.
For text extraction from publications, there are
numerous libraries in different environments, but
for brevity we only cover the most recent ones.
PDFminer.six (PDFminer, ) is a Python library capa-
ble of extracting text from PDFs and it is the most
used module for text extraction. Tika (Contributors,
b) and PDFBox (Contributors, a) are Java libraries -
also available in Python - are suitable for text extrac-
tion. Slate (Slate, 2022) is a Python library, that sup-
ports text extraction by the use of PDFminer. Textract
(Textract, 2023) is also a Python library that extracts
text from different file formats.
At the time of writing there exist only few
packages suitable for image extraction. In addi-
tion to metadata and text extraction, PDFminer.six
(PDFminer, ) also offers image extraction. PyMuPDF
DAF: Data Acquisition Framework to Support Information Extraction from Scientific Publications
469

(PyMuPDF, 2023), a Python library that extracts im-
ages from PDFs using coordinates and provides out-
put in PNG format.
There are many approaches available that extract
tables from scientific publications. Among them,
two stand out. Tabula (Tabula, 2023), a Java library
also available in Python that provides an interface for
the table extraction by pages and also supports co-
ordinate base access. Camelot (Camelot, 2023) is a
Python library capable of extracting tables from sci-
entific publications. It provides two different extrac-
tion approaches, i.e. lattice and stream, which is use-
ful for different table layouts. Moreover, it supports
extraction by coordinates and provides output in CSV
format and have been used in studies recently (Pe
˜
na
et al., 2023). Similarly it is the best performing table
extraction among several packages (Mehta, 2019).
2.2 Extraction Frameworks
By focusing on data extraction from scientific pub-
lications, numerous frameworks have emerged to
tackle the challenge of transforming unstructured
content into structured data, facilitating subsequent
tasks. These frameworks range from extracting bibli-
ographic details to capturing text, tables, and special-
ized domain-specific information. Here for simplicity
the approaches could be categorized for true digital
and scanned documents respectively.
GROBID is an open-source machine learning
library in Java for extracting, parsing, and re-
structuring raw documents into structured XML/TEI
encoded documents based on conditional random
fields (CRF). The primary functionality of GROBID
is to extract bibliographic information from scientific
publications and also to be able to extract text as well
as tables. It also has various extensions i.e. grobid-
quantities and entity fishing, which performs vari-
ous NLP tasks (Lopez, 2009),(Foppiano et al., 2019),
(Entity-Fishing, 2023).
Chemdataextractor (Swain and Cole, 2016) is a
framework, that aims for publications from the chemi-
cal domain. It captures relevant information from var-
ious data components of scientific publications and
PDFminer.six was utilised for the text extraction pro-
cess. Subsequently to enhance the extraction pro-
cess PDFDataExtractor was proposed (Zhu and Cole,
2022), it targets various aspects of information extrac-
tion considering scientific publications. PDFDataEx-
tractor for text extraction also uses PDFminer.six and
for metadata extraction they use a rule based ap-
proach considering a set of templates from domain-
specific journals. The output of the framework is sup-
posed to be passed to Chemdataextractor for relevant
Figure 1: Block Diagram of DAF Framework.
NLP tasks. Recently, in Marine Geology domain, a
framework was proposed that extracts the plain-text
from scientific publications by using PDFminer.six
and capable of extracting measurements and relevant
oceanographic and spatial information (Suryani et al.,
2022).
Furthermore, numerous approaches specialize in
information extraction from scanned documents i.e.
receipts, forms and letters rather than true PDFs.
BROS (Hong et al., 2022) is a pre-trained language
model that extracts relevant information from images
of receipts and forms and DocFormer is a transformer
base model which aims to extract various aspects
from scanned documents i.e. Dates (Appalaraju et al.,
2021).
3 DATA ACQUISITION
FRAMEWORK (DAF)
This section presents Data Acquisition Framework
(DAF) in Figure 1. In the following, the individual
modules will be elaborated thoroughly.
3.1 Document OPTICS Component
Segmentation
In order to gain a high extraction accuracy of tables
and figures, the framework utilises the “Document
OPTICS” layout analysis approach. The document
OPTICS layout analysis provides a topological sum-
mary of a document before initiating the extraction
process. This approach of detecting data components
refers to the OPTICS (Ankerst et al., 1999) algorithm.
OPTICS (Ordering Points to Identify the Clustering
Structure) is a density based algorithm for finding
clusters in spatial data. The algorithm dynamically
takes into account the average distance of all points
distances to each other within a cluster. It can detect
clusters with different densities. The densities and
distances of the points will then be plotted as a bar
chart, which is known as the characteristic OPTICS
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
470

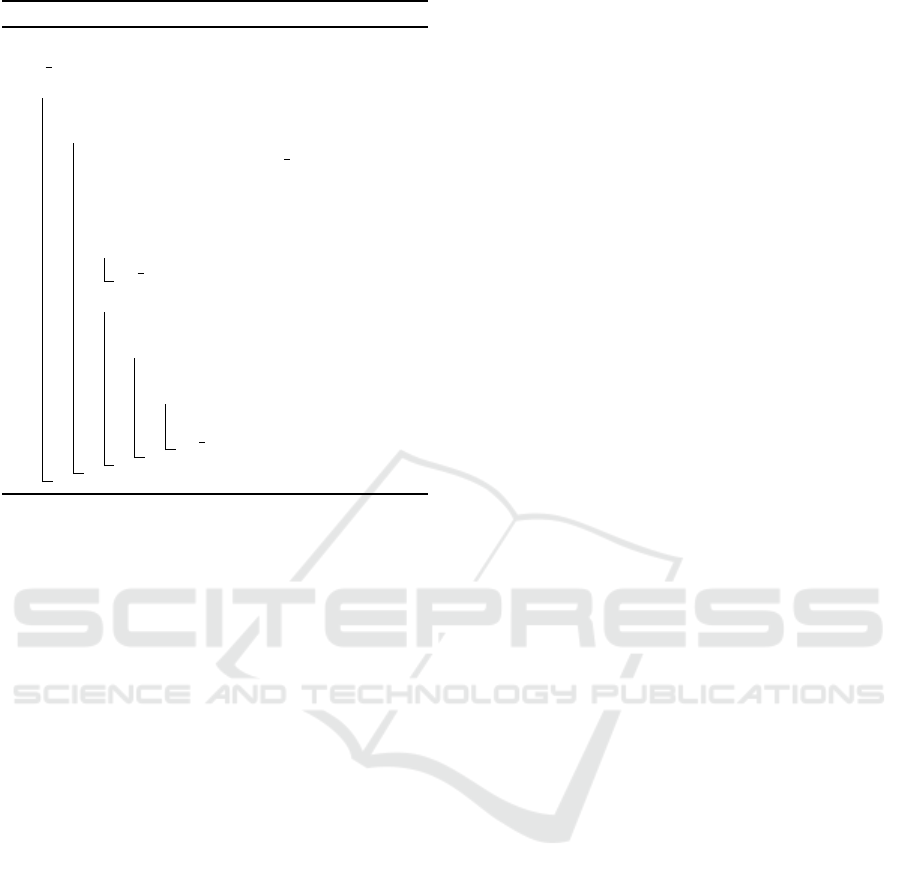
Figure 2: Number of chars per text line for extracted text of
(Kessler et al., 2006) per page.
plot. Analyzing the OPTICS plot, one can identify
data clusters. Data points, which belong to the same
cluster have on average the same height in the plot.
The same accounts for the plot of line lengths of
the extracted PDF text on Figure 2. It catches the eye
that there are regions on Figure 2 with a low aver-
age line length and as well regions with an on average
higher line length, which could be a cluster consider-
ing OPTICS. These regions, which on average share
the same height, belong to the same data component
in the publication. For lines with a low height this will
most likely be a table, because the table columns text
is extracted row by row from the PDF file.
Moreover, Figure 2 showcases the document OP-
TICS of a publication (Kessler et al., 2006). Several
components are marked with red ovals. The first one
is the abstract, since it is at the beginning of the doc-
ument and has an average line length of 100 chars per
line and second is a table caption, as it spreads over
two columns, like the most captions do and it is fol-
lowed by several short lines. The short lines marked
with a red oval for number three is text belonging to
a table, which is extracted column by column, row by
row. The fourth one is double column text: it has on
average 65 chars per line and final one indicates refer-
ences, which have on average plus ten higher number
of chars per line than the plain-text.
Using this characteristic of the publication text,
text lines can be grouped in to component and non-
component lines. This text segmentation is later used
for component extraction and PDF layout analyzer of
PDFminer (Shinyama, 2013) was used to extract the
coordinates of the horizontal text lines in the docu-
ment and horizontal text lines and their corresponding
coordinates extraction is called “coordinates file”. A
snippet of the coordinates file is shown in Figure 3.
As described earlier, the document OPTICS lay-
out analysis is used for text segmentation where text
is segmented into different groups/components. The
coordinates file is segmented into components by ap-
plying the document OPTICS to the coordinates file’s
“text” column. The result is a list of lists, where ev-
ery list includes text lines, their corresponding coor-
Figure 3: Actual Paper and Coordinate view of Table
(Riedinger et al., 2005).
dinates and indices respectively.
3.2 Caption Extraction
In scientific publications, captions an essential ele-
ment to support the extraction and indexing of figures,
tables and information linking between various data
components. The caption extraction flow is illustrated
and the detailed routine is presented in Algorithm 1.
As captions are extracted sequentially page by
page. First the publication text is split in to pages
and each page is divided into paragraphs (text blocks).
Depending on the caption layout different extraction
approaches may apply. For horizontal captions, if any
text block’s first line matches a figure/table caption it
is afterwards checked if it was matched at the begin-
ning of the line to minimize false positives. False pos-
itives occur through inlined figure/table references.
Considering vertical captions, all vertically depicted
words in the PDF will result in text blocks whose
lines are all of length one. Given the former it is first
checked whether all lines are of length one. If so, the
block is joined, reversed and checked if it is equal to
“Table” or “Figure” from line nine to eleven. When
this case applies, this match is joined with the before
block and checked if it matches the rules for caption
layout. Afterwards up to 20 text blocks (caption text)
can be appended. The result of the extraction is a
JSON file carrying captions for every page.
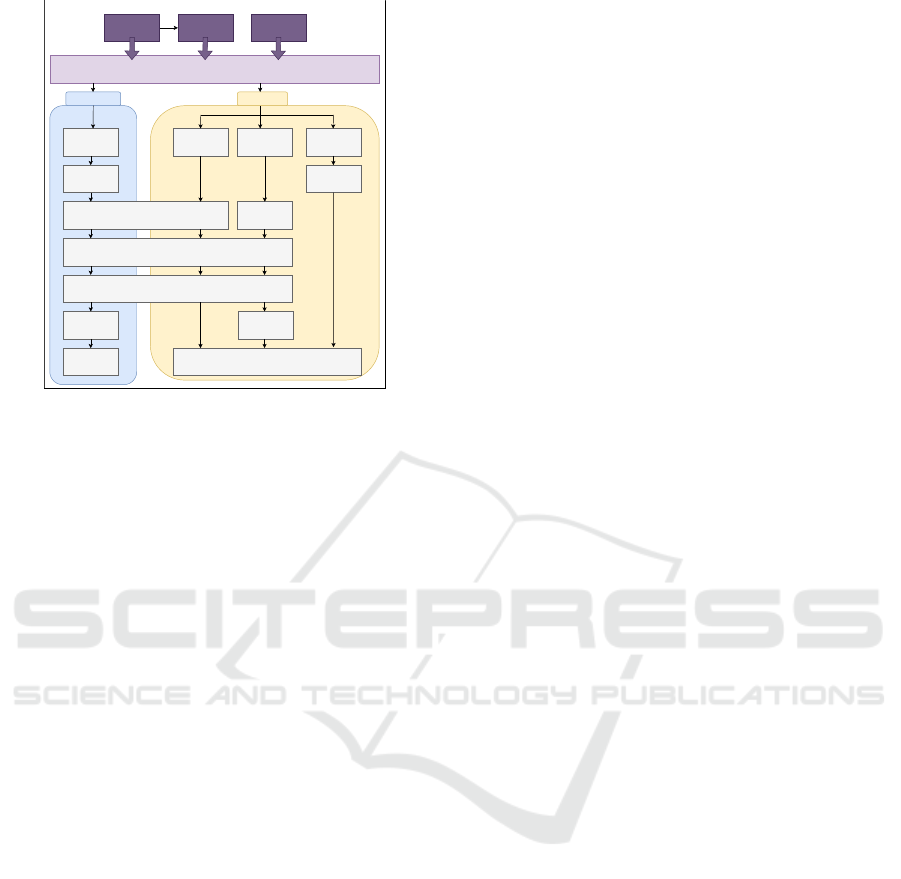
3.3 Figure and Table Extraction
The overall process of table and figure extraction is
showcased in Figure 4. Both modules are based upon
Document OPTICS text segmentation prior applied to
the coordinates file and the captions extracted from
the plain-text. The idea behind the extraction of fig-
ures and tables is to extract the top left and bottom
right coordinates from the components text lines. The
coordinates equal the maximum plane of the compo-
nent inside the PDF and can afterwards passed to the
extraction mechanism.
DAF: Data Acquisition Framework to Support Information Extraction from Scientific Publications
471
Algorithm 1: Caption Extraction.
Input : Text t
1 ft captions, pages =
/
0, t.splitByPage()
2 foreach page ∈ pages do
3 P = page.splitToParagraphs()
4 for i ∈ {0 . . . |P | − 1} do
5 cm = lambda x: caption match(x)
// lambda x:returns python re
match object
6 if cm(P
i
) ∧ cm(P
i
).startPosition() ==
0 then
7 ft captions.add(P
i
)
8 if all(map(len(line)=1,P
i
)) then
9 cap
kw
= reverse(join(P
i
))
10 if cap
kw
∈ {Table, Figure} then
11 cap
num
= P
i−1
12 if cm(cap
kw
+ cap
num
) then
13 caption = Caption(P
i
)
14 ft captions.add(caption)
3.3.1 Tables
For the extraction, the coordinates file, captions and
the PDF file is required. The coordinates file’s text
lines are segmented in to tables by utilizing the OP-
TICS layout analysis and regular expressions to fil-
ter noise. Afterwards, every page’s caption is evalu-
ated for horizontal or vertical table captions. For ev-
ery horizontal caption, the caption is searched in the
coordinates file to retrieve its starting index. In this
case the starting index is the top left coordinate of the
table and can be returned from the coordinates file.
As well, the starting index allows to select the correct
table from the segmented tables.
For table captions, there accounts the layout rule,
that a table caption must always be written on top of
the table. Therefore the starting index of the first table
caption line must be smaller than the first line of the
table group and higher than the last line index of the
prior table group. After selecting the correct group, in
order to extract the table with camelot by coordinates
xy1 and xy2 coordinates have to be retrieved. The
top left coordinates can be taken from the caption as
mentioned earlier. The bottom right coordinates can
be retrieved from searching the maximum x2 and y2
coordinates from the selected table group. These co-
ordinates are then passed to camelot module, which
extracts the table in CSV format.
For every vertical table caption it is important to
mention again, that vertical table’s text lines are all
of length one. Therefore it is not possible to find the
caption’s starting index as it is done for the horizon-
tal captions. First all white-space is removed from the
caption. If one text line equals the letter “T” and its
predecessors equal the rest of the caption the starting
index of the caption is found and likewise the xy1 co-
ordinates. With the starting index, the table group is
selected and from the table group the maximum span
is retrieved. Later on, the page is rotated by 90 de-
grees and the coordinates are passed to camelot mod-
ule for extraction.
3.3.2 Figures
In general there are mostly two types of figures
included in a scientific publication: Portable Net-
work Graphic (PNG) or graphical based formats
(PDF/SVG). Regarding graphical based formats, the
xy-ticks of a diagram or plot can be extracted by
PDFminer. In the document OPTICS plot those ticks
appear as a region with on average low line lengths
and could be wrongly identified as a table. Just as in
the table extraction module, every figure’s caption in-
dex is retrieved from the coordinates file in order to
use it for the group selection. Generally, figure cap-
tions are located below the figure. For the group se-
lection the figure caption index must be greater than
the last group’s line index and smaller than the first
line of the next group’s index. Later on, the maxi-
mum xy-span is selected from the group in the same
way as retrieving the maximum span from the table
group.
In order to be able to extract graphical based fig-
ures, all PDF pages need to be converted to PNGs.
Afterwards the xy-span of the PDF images and the
coordinates file is obtained. This is necessary to con-
vert the coordinates obtained from the figure group
(PDFminer coordinates) to PNG coordinates. Along
with the converted coordinates the graphical based
image can be cut out from the original PDF page, that
was converted to PNG.
3.4 Section Extraction
The section extraction module extracts the text of the
publications segmented in to its section structure. Ini-
tially, the section modules gets passed the plain-text,
where all lines non-text-lines (figures, tables, etc.)
have been removed by the prior modules. The next
step is to parse the text for its sections. Essential for
the extraction accuracy is the correct parsing for sec-
tion titles. This is done by regular expressions. The
routine iterates over all text paragraphs and checks if
the first line matches a section title. If this is the case,
the title is inserted in to a dictionary. Otherwise, the
text of the paragraph is appended to the values of the
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
472
CaptionsCoordinates File
OPTICS
Line Grouping
For All Page's Captions
Convert
PDF to Images
Vertical
get xy-span from
1. pdf-images
2. coords.-file
No Table CaptionHorizontal
Find Caption Line Index in Coords.-File
Lookbehind Search
Line Index Selection
Group Selection by Caption Line Index and Component Style
Get Maximum xy-span from group
Coordinate
Transformation
Rotate Page
by 90°
Camelot
Open-CV
Cut Out Image by
Coordinates
OPTICS
Table Detection
Figures Tables
Figure 4: Table and Figure Extraction Flow Diagram.
last key inserted. Moreover, auxiliary sections like
”Appendix” or ”Acknowledgements” are appended in
section dictionary. Likewise the section’s sentences
are pre-processed, cleaned and matched for certain
patterns defined by regular expressions. Making use
of the NLP library spaCy (Honnibal et al., 2020), the
text is parsed into sentences. Finally the abstract and
references are inserted at their respective positions in
the dictionary.
3.5 Metadata Extraction
This section briefly explains the metadata extraction
process of Data Acquisition Framework. Generally,
metadata features can be located on the first page of a
publication. The following list provides an overview:
• DOI: The framework first looks for the DOI in the
metadata file generated by the Tika module, or, if
not found, it tries to locate the DOI within any line
on the first page using a regular expression.
• Citation/Dates: both can also be found by sub-
string match for ”citation”, ”received” or ”ac-
cepted” keywords.
• Affiliations can be found by searching variations
of ”University of” in different languages with a
regular expression.
• Abstract For abstracts, the text is divided at the
position of ”Abstract” keyword and afterwards
again it is divided at the position of ”Introduc-
tion” keyword and the text between the keywords
is separated. From the given data it is experienced,
that the average length of abstract ranges from 300
to 600 words. Each paragraph is added to the
string, which will in the end be the abstract, while
it is not longer than 600 words. The word limit
is especially helpful for abstracts, which are not
written in a single paragraph. Therefore all the ab-
stract layouts are covered. Regarding the text line
length condition, the most abstracts’ length span
over two columns and layouts can differ. There-
fore only if two-thirds of the text blocks lines are
longer than 80 characters, then the text block is
accounted as an abstract paragraph. For abstracts,
which are written in single column layout the char
limit is 65 characters per line.
• Title: First the framework tries to extract it from
the Tika metadata file, otherwise searches it on the
first page of publication. The title is mostly writ-
ten on the top of the first page after journal name
and citation, but before the authors, abstract and
keywords. In most cases it is the only paragraph
left after everything else was separated.
4 EXPERIMENTAL RESULTS
This section presents a comprehensive experimental
evaluation of the Data Acquisition Framework.
4.1 Dataset
For the experimental evaluation of DAF, we con-
sidered two distinct dataset. Firstly, we replicated
the dataset used in evaluating the PDFDataExtractor
framework from the Chemical Domain, which com-
prises of 100 full publications collected from five
chemical domain journals. The second dataset was
gathered from the field of Marine Science comprises
of 700 full papers from various journals, maintaining
the same criteria as the PDFDataExtractor module i.e.
Elsevier Journal Publications only. It is important to
note that PDFDataExtractor strongly relies on tem-
plates, unlike our proposed framework (Zhu and Cole,
2022). Hence, to highlight DAF’s flexibility, we also
processed research reports from Marine Science ex-
peditions.
4.2 Chemical Domain Results
In this regard, initially we replicated the results col-
lected by PDFDataExtractor on chemical domain
publications and process similar papers using our
module. For the sake of generality, we adhered to
the evaluation criteria defined by the PDFDataEx-
tractor framework (Zhu and Cole, 2022). Typically,
the evaluation process is manual, making it a semi-
manual process overall. The results for the metadata
DAF: Data Acquisition Framework to Support Information Extraction from Scientific Publications
473
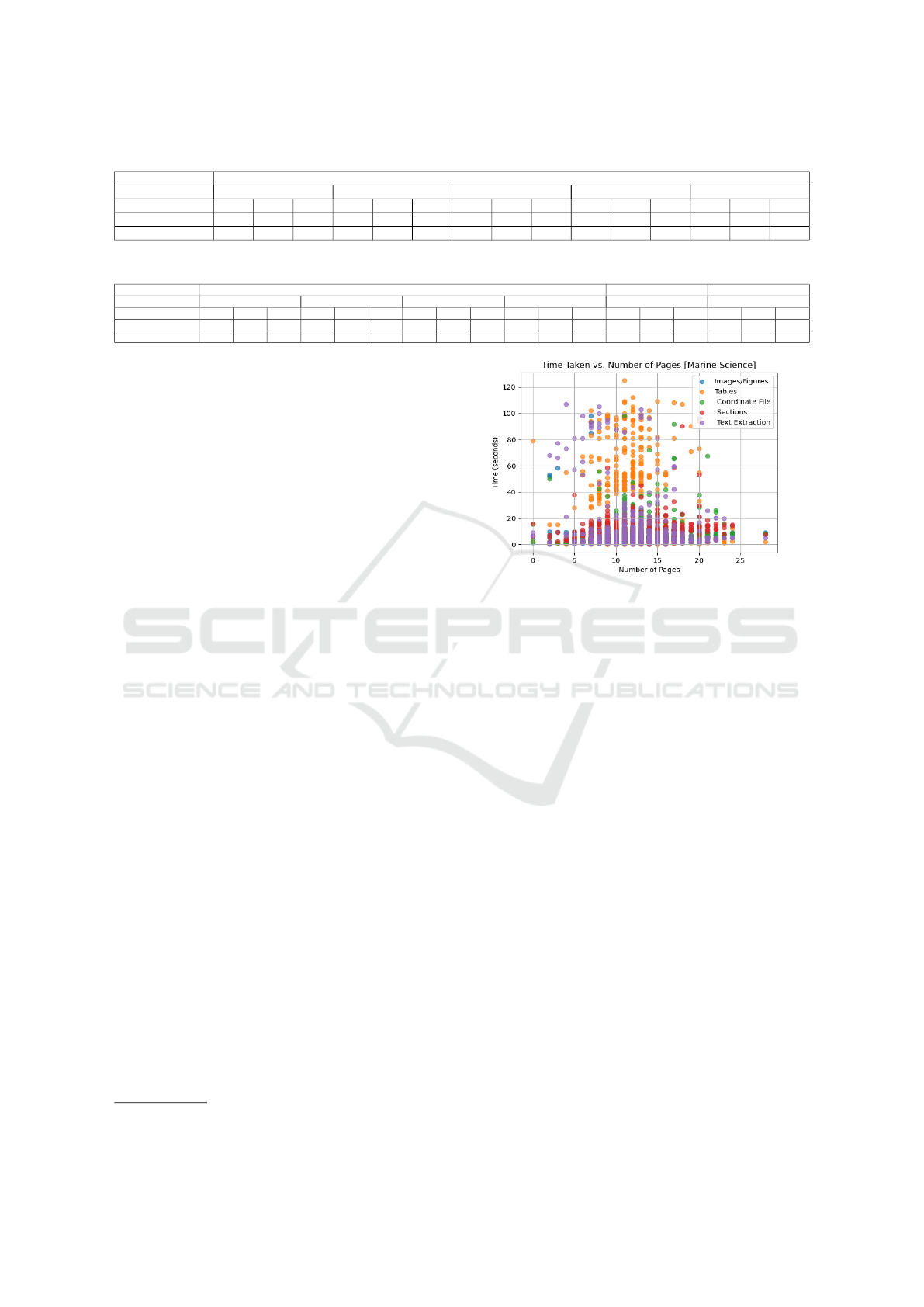
Table 1: Results Chemical Domain Publications (Metadata).
Framework Metadata
DOI Title Abstract Authors Journal
P R F1 P R F1 P R F1 P R F1 P R F1
PDFDataExtractor 0.928 0.966 0.944 0.925 0.944 0.932 0.854 0.862 0.852 0.466 0.527 0.480 0.244 0.528 0.304
DAF 1.0 0.950 0.974 1.0 0.950 0.975 0.932 0.588 0.715 1.0 0.916 0.956 1.0 0.950 0.974
Table 2: Results of Chemical Domain Publications (Paper Content).
Framework Captions and Content Body References
Figure Caption Figure Content Table Caption Table Content Sections References
P R F1 P R F1 P R F1 P R F1 P R F1 P R F1
PDFDataExtractor 0.811 0.840 0,818 - - - - - - - - - 0.650 0.905 0.746 0.566 0.694 0.588
DAF 0.952 0.927 0.939 0.484 0.948 0.641 0.969 0.969 0.969 0.839 0.894 0.866 0.968 0.634 0.766 1.0 0.690 0.816
extraction task is compiled in Table 1 indicating that
DAF performs well overall. Table 2 presents the re-
sults collected considering the content of the papers.
Hence, DAF is able to cover a broader spectrum of
data components from publications in comparison to
PDFDataExtractor, The “-” signs in tables indicates
that the system was unable to extract the respective
content. The final results indicate that our framework
perform better in comparison to the baseline.
During the evaluation, it has been experienced that
PDFDataExtractor is unable to process a good set of
papers. In contrast, DAF was able to successfully pro-
cess all the provided files. For simplicity , we abbre-
viated Precision, Recall, and F1-Score as P, R, and F1,
respectively in corresponding tables.
4.3 Marine Science Results
Similarly, papers from the Marine Science domain
is processed using both the PDFDataExtractor and
DAF. The outcomes of the metadata extraction are
presented in 3, which emphasises the effectiveness
of our proposed framework. Moreover, Table 4 high-
lighted the results for the content of the papers. For
tables evaluation we followed: headers of the tables
and data in columns remain intact and no rows were
lost. Additionally, tables in Marine Science papers
typically contain diverse numerical expressions and
spatial data.
4.4 IODP Results
International Ocean Discovery Program (IODP) is
global scientific research program that conducts expe-
ditions at various oceanographic locations and reports
of these expeditions are available in their publication
portal
1
. These reports are in PDFs, but not follow the
standard publication formats. Generally, these report
comprise of text, tables, figures. So, we processed
these reports with both frameworks. DAF was able
1
http://publications.iodp.org
Figure 5: Number of Pages and Time plot.
to extract the IDs, titles, authors, source, table cap-
tions and table content respectively, but PDFDataEx-
tractor framework was unable to process even a single
report. The gathered results are presented in Table 5,
indicating the DAF’s ability to process PDF files in
general with fewer template dependency in compari-
son to PDFDataExtractor.
4.5 Overall Results
This sections discusses the overall performance of
DAF. We presented the response time of DAF for in-
dividual components with respect to number of pages
shown in Figure 5. For table detection, we evaluated
our coordinate based approach to base camelot mod-
ule on Marine Science publications and results are
presented in Figure 6. Our approach detects 1241 ta-
ble instances out of 700 files, which is inline with the
actual number of 1255 and base camelot detects 4782
instances. For Chemical domain publications, there
are 144 tables are in actual and our approach detect
128 table instances, beside, base camelot module de-
tects 440 table instances.
4.6 Information Modelling
To demonstrate efficacy of DAF, we presented a com-
plex use-case which could facilitate research data
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
474

Table 3: Results of Marine Science Publications (Metadata).
Framework Metadata
DOI Title Abstract Authors Journal
P R F1 P R F1 P R F1 P R F1 P R F1
PDFDataExtractor 1.0 0.958 0.978 0.924 0.923 0.923 0.931 0.506 0.656 0.937 0.253 0.399 0.965 0.547 0.698
DAF 0.996 0.988 0.992 1.0 1.0 1.0 0.917 0.930 0.923 0.999 0.975 0.987 1.0 0.984 0.992
Table 4: Results of Marine Science Publications (Paper Content).
Framework Captions and Content Body References
Figure Caption Figure Content Table Caption Table Content Sections References
P R F1 P R F1 P R F1 P R F1 P R F1 P R F1
PDFDataExtractor 0.972 0.936 0.954 - - - - - - - - - 0.883 0.865 0.874 0.321 0.127 0.182
DAF 0.980 0.930 0.954 0.994 0.829 0.903 0.997 0.980 0.988 0.921 0.945 0.933 0.997 0.674 0.804 0.997 0.977 0.987
Table 5: Results of IODP Expedition Reports.
Framework Metadata Caption and Content
E-DOI Title Authors Source Table Caption Table Content
P R F1 P R F1 P R F1 P R F1 P R F1 P R F1
PDFDataExtractor - - - - - - - - - - - - - - - - - -
DAF 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 0.985 1.0 0.993 0.730 0.902 0.807
Figure 6: Table Detection for Marine Science.
management. We segregated a set of publications
from Marine Science carrying spatial coordinates in
tables i.e. latitude and longitude. These coordinates
are being represented in degrees/decimals and could
be a perfect case to demonstrate the usefulness of ex-
tracted information. Out of 700 files 73 publications
are carrying spatial coordinates in tables. Among
these tables, there are total 2092 location instance
were observed and 1855 of them are the expressions
that could lead to true spatial coordinates. In this ex-
ercise, 237 location instances were missed/extraction
anomalies, which rather indicates encoding issues.
For example: degree sign were converted to “0” / “8”
and minute sign was converted to “1”.
5 CONCLUSIONS
The Data Acquisition Framework (DAF) significantly
broadens the scope of information extraction by en-
compassing more data components from scientific
publications by showcasing its potential to adhere di-
verse information extraction in comparison to base-
line. Its hybrid approach to tackle metadata proved
successful as indicated by the gathered results. For
extracting captions, tables and sections, DAF also
highlights its authority, leveraging document OPTICS
and coordinate files. However, there is room for en-
hancement in figure extraction, currently the system
is not able to group multiple figures under a single
caption. Experimental results have demonstrated that
DAF exhibits minimal template dependency and ex-
cels in precise data extraction from various compo-
nents as revealed in information modelling.
Moreover, the possible enhancements in DAF may
include precise caption-to-image mapping, challenge
of associating non-graphical figures with their respec-
tive captions. Additionally, the framework could in-
clude the extraction of mathematical formulas, pre-
serving their integrity during extraction and conver-
sion to LaTeX code. Such enhancements would be
particularly valuable in applications related to the
natural sciences. Furthermore, DAF presents excit-
ing opportunities for future research. It could serve
as a candidate for creating an image data repository
by extracting images from publications. Addition-
ally, the extracted data from tables could contribute
to the development of a Knowledge Graphs, Recom-
mender Systems for scientific publications, encom-
passing both metadata and paper content. Hence, the
publications in this regard are a horizon of informa-
tion, DAF could be a step towards extraction of rele-
vant information for the knowledge discovery and re-
search data management tasks.
DAF: Data Acquisition Framework to Support Information Extraction from Scientific Publications
475

ACKNOWLEDGEMENTS
This work was supported by the Helmholtz School for
Marine Data Science (MarDATA) partially funded by
the Helmholtz Association (grant HIDSS-0005).
REFERENCES
Ankerst, M., Breunig, M. M., Kriegel, H.-P., and Sander, J.
(1999). Optics: Ordering points to identify the clus-
tering structure. ACM Sigmod record, 28(2):49–60.
Appalaraju, S., Jasani, B., Kota, B. U., Xie, Y., and Man-
matha, R. (2021). Docformer: End-to-end transformer
for document understanding. In Proceedings of the
IEEE/CVF international conference on computer vi-
sion, pages 993–1003.
Camelot (2023). camelot. [Online; accessed: March 21,
2023].
Chamberlain, S. (2019). fulltext: Full text of ’scholarly’
articles across many data sources. R package version
1.4.0.
Chamberlain, S., Zhu, H., Jahn, N., Boettiger, C., and
Ram, K. (2023). rcrossref: Client for various
’crossref’ ’apis’. https://docs.ropensci.org/rcrossref/,
https://github.com/ropensci/rcrossref.
Contributors, G. Apache pdfbox - a java pdf library. https:
//pdfbox.apache.org/. [Online; accessed: February 20,
2023].
Contributors, G. Apache tika - a content analysis toolkit.
https://tika.apache.org/. [Online; accessed: February
20, 2023].
Entity-Fishing (2016–2023). entity-fishing. https://github.
com/kermitt2/entity-fishing.
Foppiano, L., Romary, L., Ishii, M., and Tanifuji, M.
(2019). Automatic identification and normalisation of
physical measurements in scientific literature. In Pro-
ceedings of the ACM Symposium on Document Engi-
neering 2019, pages 1–4.
Guerra, J., Quan, W., Li, K., Ahumada, L., Winston, F.,
and Desai, B. (2018). Scosy: A biomedical collabo-
ration recommendation system. In 2018 40th annual
international conference of the IEEE engineering in
medicine and biology society (EMBC), pages 3987–
3990. IEEE.
Hong, T., Kim, D., Ji, M., Hwang, W., Nam, D., and
Park, S. (2022). Bros: A pre-trained language model
focusing on text and layout for better key informa-
tion extraction from documents. In Proceedings of
the AAAI Conference on Artificial Intelligence, vol-
ume 36, pages 10767–10775.
Honnibal, M., Montani, I., Van Landeghem, S., and Boyd,
A. (2020). spaCy: Industrial-strength Natural Lan-
guage Processing in Python.
Inc., A. S. (2006). PDF Reference Version 1.7. 6 edition.
Jinha, A. E. (2010). Article 50 million: an estimate of the
number of scholarly articles in existence. Learned
publishing, 23(3):258–263.
Kessler, J., Reeburgh, W., Southon, J., Seifert, R.,
Michaelis, W., and Tyler, S. (2006). Basin-wide
estimates of the input of methane from seeps and
clathrates to the black sea. Earth and Planetary Sci-
ence Letters, 243(3-4):366–375.
Kreutz, C. K. and Schenkel, R. (2022). Scientific paper
recommendation systems: a literature review of re-
cent publications. International Journal on Digital
Libraries, 23(4):335–369.
Lopez, P. (2009). Grobid: Combining automatic biblio-
graphic data recognition and term extraction for schol-
arship publications. In Agosti, M., Borbinha, J., Kap-
idakis, S., Papatheodorou, C., and Tsakonas, G., ed-
itors, Research and Advanced Technology for Dig-
ital Libraries, pages 473–474, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Martinez-Rodriguez, J. L., Hogan, A., and Lopez-Arevalo,
I. (2020). Information extraction meets the semantic
web: a survey. Semantic Web, 11(2):255–335.
Mehta, V. (2019). Comparison with other pdf table extrac-
tion libraries and tools.
PDFminer. pdfminer.six. https://github.com/pdfminer/
pdfminer.six. [Online; accessed: December 20, 2022].
Pe
˜
na, A., Morales, A., Fierrez, J., Ortega-Garcia, J.,
Grande, M., Puente, I., Cordova, J., and Cordova, G.
(2023). Document layout annotation: Database and
benchmark in the domain of public affairs. arXiv
preprint arXiv:2306.10046.
PyMuPDF (2023). Pymupdf. [Online; accessed: March 21,
2023].
Riedinger, N., Pfeifer, K., Kasten, S., Garming, J. F. L.,
Vogt, C., and Hensen, C. (2005). Diagenetic alter-
ation of magnetic signals by anaerobic oxidation of
methane related to a change in sedimentation rate.
Geochimica et Cosmochimica Acta, 69(16):4117–
4126.
Shinyama, Y. (2013). Programming with pdfminer.
Slate (2022). Slate. [Online; accessed: November 07,
2022].
Suryani, M. A., Wolker, Y., Sharma, D., Beth, C., Wall-
mann, K., and Renz, M. (2022). A framework for ex-
tracting scientific measurements and geo-spatial infor-
mation from scientific literature. In 2022 IEEE 18th
International Conference on e-Science (e-Science),
pages 236–245. IEEE.
Swain, M. C. and Cole, J. M. (2016). Chemdataextractor: a
toolkit for automated extraction of chemical informa-
tion from the scientific literature. Journal of chemical
information and modeling, 56(10):1894–1904.
Tabula (2023). Tabula. [Online; accessed: February 15,
2023].
Taylor-Sakyi, K. (2016). Big data: Understanding big data.
arXiv preprint arXiv:1601.04602.
Textract (2023). Textract. [Online; accessed: March 21,
2023].
Yadav, P., Remala, N., and Pervin, N. (2019). Reccite: A
hybrid approach to recommend potential papers. In
2019 IEEE international conference on big data (big
data), pages 2956–2964. IEEE.
Zhu, M. and Cole, J. M. (2022). Pdfdataextractor: A tool
for reading scientific text and interpreting metadata
from the typeset literature in the portable document
format. Journal of Chemical Information and Model-
ing, 62(7):1633–1643.
KDIR 2023 - 15th International Conference on Knowledge Discovery and Information Retrieval
476
