
A WEB-BASED ARCHITECTURE FOR INDUCTIVE LOGIC
PROGRAMMING IN BIOLOGY
Andrei Doncescu
Laas, Toulouse, France
Katsumi Inoue
National Institute for Informatics, Tokyo, Japan
Muhammad Farmer, Gilles Richard
British Institute for Technology and E-commerce, London, UK
Keywords: Internet services, Inductive Logic Programming, biological application
Abstract: In this paper, we present a current cooperative work involving different institutes around the world. Our aim
is to provide
an online Inductive Logic Programming tool. This is the first step in a more complete structure
for enabling e-technology for machine learning and bio-informatics. We describe the main architecture of
the project and how the data will be formatted for being sent to the ILP machinery. We focus on a biological
application (yeast fermentation process) due to its importance for high added value end products.
1 INTRODUCTION
Our main aim is to provide a web-based machine
learning application. On one hand, we have at our
disposal a powerful inductive machinery
(inoue2004), enjoying interesting theoretical
properties (soundness, completeness) and whose
current version is implemented in JAVA. On the
other hand, there is a considerable effort made by
the biologists to understand and then to control a
well known bioprocess, namely yeast fermentation.
None of the available mathematical models (mainly
based on dynamic differential equations) allows a
precise understanding. But the possibility to
understand the biological processes leading to high
added value end-products like vitamins,
antibiotics,… is highly challenging. For instance, it
has recently been shown that the human yeast carry
out synthetically the different tasks of producing the
human biosynthesis protein. The recent results show
that a "humanised yeast" could simplify drug
manufacture by introducing a human gene inside
yeast chromosome. Since yeast grow faster and need
less tending than mammalian cells, it could be a
solution to produce proteins cheaper and easier.
More than that, there is today a huge effort to
imagine alternative to classical petrol-based fuel.
Ethanol is one of the possible solution. Several
governments encourage biologists to focus on bio-
processes leading to ethanol as side product of a
whole process. The more the final percentage of
ethanol, the more the fermentation process is
successful. It makes no doubt that, in a sustainable
development perspective, this research topics has to
be more deeply investigated. But, as previously
explained, it is difficult to produce these proteins to
a commercial scale due to the incapacity of the
biologist to keep the cell on a specific pathway. Due
to the lack of relevant models, micro-biologists are
not able to dynamically tune the relevant parameters
insuring a high percentage of ethanol output.
In this project, our aim is to apply techniques
issued from the field of In
ductive Logic
Programming to complement the standard
mathematical tools. We hope that our inductive
machinery will be able to highlight the relevant
parameters, and more than that, to provide simple
explanations in a logical form. Not only, we want to
send the output of the yeast fermentation observation
to an inductive machine, but we also focus on the
possibility to send online the data to the ILP
357
Doncescu A., Inoue K., Farmer M. and Richard G. (2005).
A WEB-BASED ARCHITECTURE FOR INDUCTIVE LOGIC PROGRAMMING IN BIOLOGY.
In Proceedings of the Seventh International Conference on Enterprise Information Systems, pages 357-361
DOI: 10.5220/0002516903570361
Copyright
c
SciTePress

machine. In this paper, we investigate the structure
of our web-based application.
The next section is devoted to a general presentation
of the advantages e-technology can bring in the
context of machine learning and especially in bio-
informatics. Section 3 briefly recall the main
philosophy of inductive logic programming, as a
sub-topic of machine learning. In section 4, we
describe how we process the data, using XML as a
backbone format, and we present the general
architecture of our system. We conclude in section5.
2 E-TECHNOLOGY
E- Technology has made it possible to carry out lot
of activities, previously locally achieved and are
remotely performed. All these activities constitute
what is usually called E-business. Government and
organisation have come to realise the value of e-
technology hence such projects i.e. e-learning, e-
government, e-commerce, e-health are being
implemented globally. In some sense, bio-
informatics aims to solve biological problems using
computing methods. That is why we think it is
essential to make bio-informatics benefit of these
techniques because the bio-informatics definitions
are changing in accordance with the development of
new areas in science. BITE (UK), IRIT (France),
LAAS (France) and NII (Japan) focus together on
maturing technology to globalise development and
research of bio-informatics.
A global platform [farmer and al.2004],
integrating grid computing, programming tools and
data visualization technologies would enable
scientists to gain greater insight into their research
through direct comparisons of simulations,
experiments and observations. Knowledge
repositories maintaining relationships, concepts, and
inference rules would be accessible through such a
global platform. Starting from these ideas, we
develop a web-based architecture around a machine
learning application : the target machinery is an
Inductive Logic Programming (ILP) system and the
target application is a bio-process leading to ethanol.
The next sections are devoted to a more precise
investigation of these two fields.
But it should be clear that our scheme is open
and that there is no conceptual obstacle to integrate
other inductive engines as well as there is no
obstacle to focus on other fields than bio-informatics
Starting from these ideas, we develop a web-
based architecture around a machine learning
application : the target machinery is an Inductive
Logic Programming (ILP) system and the target
application is a bio-process leading to ethanol. The
next sections are devoted to a more precise
investigation of these two fields.
But it should be clear that our scheme is open and
that there is no conceptual obstacle to integrate other
inductive engines as well as there is no obstacle to
focus on other fields than bio-informatics.
3 INDUCTIVE LOGIC
PROGRAMMING: A BRIEF
REVIEW
Inductive Logic Programming (ILP) is the part of
machine learning where the underlying model is
described in term of first-order logic. We briefly
outline here the standard definitions and notations.
Given a first-order language L with a set of variables
Var, we build the set of terms Term, atoms Atom and
formulas as usual. The set of ground terms is the
Herbrand universe H and the set of ground atoms or
facts is the Herbrand base B. A literal l is just an
atom a (positive literal) or its negation ~a (negative
literal). A substitution s is an application from Var
to Term with inductive extension to Atom. We
denote Subst the set of ground substitutions.
A clause is a finite disjunction of literals and a
Horn clause is a clause with at most one positive
literal. A Herbrand interpretation I is just a subset of
B : I is the set of true ground atomic formulas and its
complementary denotes the set of false ground
atomic formulas.
We can now proceed with the notion of logical
consequence. Given A an atomic formula, I, s |- A
means that s(A) belongs to I. As usual, the
extension to general formulas F uses
compositionality.
• I |- F means : forall s, I, s |- F (we say I is
a model of F).
• |- F means : forall I, I |- F.
• F |- G means that all models of F are
models of G.
Stated in the general context of first-order logic,
the task of induction is to find a set of formulas H
such that:
B U H |- E
Given a background theory B and a set of
observations E (training set), where E, B and H here
denote sets of clauses. In this paper, E is always
ICEIS 2005 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
358

given as positive examples, but negative examples
can also be introduced as well.
A set of formulas is here, as usual, considered as the
conjunction of its elements. Of course, one may add
two natural restrictions~:
• ~ (B |- E) since, in such a case, H would not
be necessary to explain E.
• ~ (B U H |-
┴
) : this means B U H is a
consistent theory.
In the setting of relational databases, inductive
logic programming (ILP) is often restricted to Horn
clauses and function-free formulas, E is just a set of
ground facts. The main reason of this restriction is
due to easiness for handling such formulas. An
extension to non-Horn clauses in B, E, and H is,
however, useful in many applications. For example,
indefinite statements can be represented by
disjunctions with more than one positive literals, and
integrity constraints are usually represented as
clauses with only negative literals. A clause-form
example is also useful to represent causality. The
inductive machinery developed by (inoue, 2004) can
handle all such extended classes.
There are two other remarks on the logic for
induction.
• The distinction between B and E is a matter
of taste. In fact, some induction problems
which often be seen in data mining do not
distinguish between B and E, and extracts
rules merely from the whole knowledge
base. However, the distinction is
important from practical viewpoints [inoue
and al2004]. When we already have our
current knowledge B and then a new
observation E is obtained to update B, this
E should be assimilated into our knowledge
in a way that E should change the current
theory B into the augmented theory B U H
such that B U H |-
E holds. In this case,
background knowledge is intrinsic to
knowledge evolution. We cannot realize
continuous and incremental learning if we
merely treat examples without any prior
knowledge.
• When we investigate induction deeper,
some subtleties appear according to the
properties of induction (e.g., whether the
closed-world assumption is applied or not).
These issues are out of the scope of this
paper. See [inoue and al. 2004].
We give an example for an induction problem
for the inductive machinery by (inoue, 2004).
Suppose B contains only two rules B1 and B2 such
that every cat is a pet (B1) and that if a pet is small
and fluffy then it is cuddly (B2):
input(b1,bg,[-cat(X), +pet(X)]).
input(b2,bg,[-small(X), -fluffy(X), -
pet(X), +cuddly(X)]).
Suppose we observe E that any fluffy cat is
cuddly:
input(e1,obs,[-fluffy(X),-cat(X),+cuddly(X)]).
We also put some control information for the
inductive machinery such that the maximum allowed
length of clauses and the maximal term depth:
production_field([length <= 3, term_depth
< 3]).
strategy(depth_first_iterative_deepning([dep
th <= 4,iterative_depth_step = 1])).
inductive_bias([include_carc = 0, lgg = 0
, dropping = 1]).
Then we get the following result as H, namely, a
fluffy cat is small:
% java CF problem/example2.ax
Observations E: [-fluffy(_X), -cat(_X),
cuddly(_X)]
Background B:
[-cat(_X), pet(_X)]
[-small(_X), -fluffy(_X), -pet(_X),
cuddly(_X)]
Hypotheses:
[[-pet(_X), -fluffy(_X), -cat(_X),
small(_X), cuddly(_X)]]
selecting dropping literals in the
clause:
1,5.
Hypotheses:
[[-fluffy(_X), -cat(_X), small(_X)]]
That is the simple implication :
cat(X) and fluffy(X) -> small(X)
This is easily readable as a natural language
sentence :
A fluffy cat is small.
This is one of the great feature of first order
logic : the translation of the output into a human-
readable format is rather straightforward.
4 THE GENERAL
ARCHITECTURE
The expected architecture is rather simple and in
accordance with the n-tier e-application model. We
begin with the data i.e. the set E we have to provide
to the machine.
A WEB-BASED ARCHITECTURE FOR INDUCTIVE LOGIC PROGRAMMING IN BIOLOGY
359
4.1 The data
We distinguish 2 kinds of parameters which are
meaningful in the whole process and for which we
have data :
a)Macroscopic parameters : temperature,
pressure, increasing rates for these parameters
(kinetics of the process), etc.. These parameters
are evaluated online with the relevant sensors.
Each measure has a very low cost, so we can
potentially dispose of a huge database. These
parameters have numerical values.
b)Microscopic parameters : we need to
distinguish here two kinds :
- Cellular level : by image analysis, we can
get the form of a cell (spheric, elliptic, etc…).
These parameters are symbolic in nature and
belong to a finite set of values.
- Molecular level : these parameters
describe the DNA structure of the target
molecule. This kind of analysis is not only very
long to be achieved but very expensive. That is
why we cannot get a lot of data from this side.
These parameters have symbolic values.
Some other parameters are available but it is not
clear today if they make sense for the whole process.
The inductive machinery supplied by ILP usually
takes inputs with symbolic representation. For this
reason, numerical values are often discretized into
Boolean values like +1 (positive/increasing), 0
(neutral/no change), -1 (negative/decreasing), by
using theresholds and the multinominal distribution.
These Boolean values are suited for modeling
qualitative behaviors between parameters. Of course,
some intermediate values can be associated as
representation of fuzziness. For more complex
domains, we need a regression method to detect
linear and nonlinear relationships. A more practical
method would employ a profile data for an
important parameter.
We understand that we have a lot of parameters
to deal with. The data are abstractly described in an
XML format. Typical DTD definition of our data is :
<!ELEMENT dataset (data*)>
<!ELEMENT data (name, type, value>
And a set of data looks like :
<dataset>
<data>
<name>pH</name>
<type>discrete</type>
<value>6.5</value>
</data>
<data>…</data>
</dataset>
Since the real data are mainly provided as a xls
format, there is no difficulty to compile into the
XML format. This XML file is sent to the server. A
simple compiler converts the XML files into
convenient data input format for the target ILP
machine. The output of the ILP machinery is just a
finite set of logical rules which are translated into a
human readable format, as previously explained.
There is few things to do since the rules are put in an
implicative form and this is generally
understandable as soon as the predicate names
support intuition.
4.2 The background knowledge
There is a very huge amount of litterature about the
subject and we have to extract, with the help of
biologists, the rules which could be useful as a way
to guide the research (see (Doncescu and al.2005)).
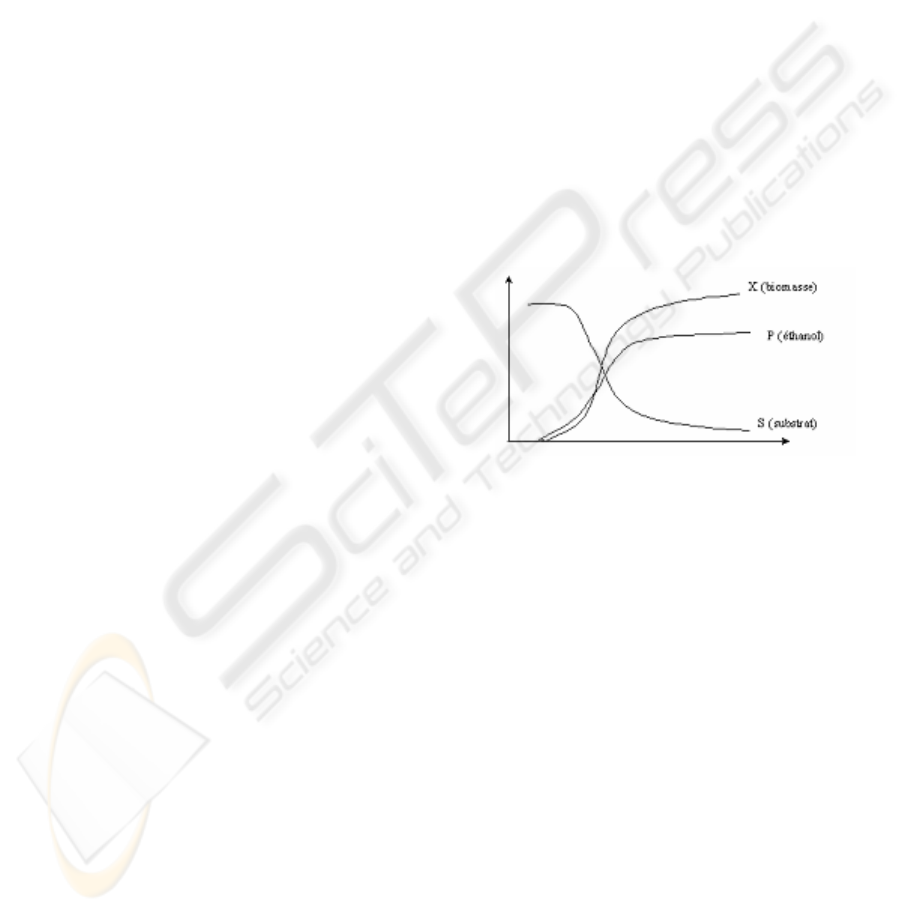
The experimental evolution for state variable of the
system : biomass, substrate and product could be
figure up by the curves below :
Figure 1
We understand that the consumption of substrat
increases the number of interesting cells (parameters
X and P (ethanol partial pressure)). This is translated
into a very simple logical rule :
not increase(S,t) OR [(increase(P,t) AND
increase(X,t))]
which is equivalent into
[not increase(S,t) OR increase(P,t)]
AND [not increase(t)
OR increase(,t)]
We get a set of 2 clauses (conjonctive normal
forms). The parameter t, representing the time,
appears in every predicate in order to capture the
dynamic of the process. The time is considered as a
discrete attribute corresponding to the discrete
sample steps. Such rules are translated in an XML
format, in order to be easily adaptable to the input
format of the remote ILP machine. It is remarkable
to see that XML format is very close to a logical
format. For instance, some DTD rules for a logic
program (not reduced to Horn clauses) are :
<!ELEMENT cnf (disjunction*)>
<!ELEMENT disjunction (literal*)>
ICEIS 2005 - ARTIFICIAL INTELLIGENCE AND DECISION SUPPORT SYSTEMS
360
<!ELEMENT literal (predicate,
argument*)>
In this case, the translation is straightforward and
there is no need to spend a lot of time for that. This
is mainly a recursive walk inside a text file.
4.3 A functional view of the whole
system
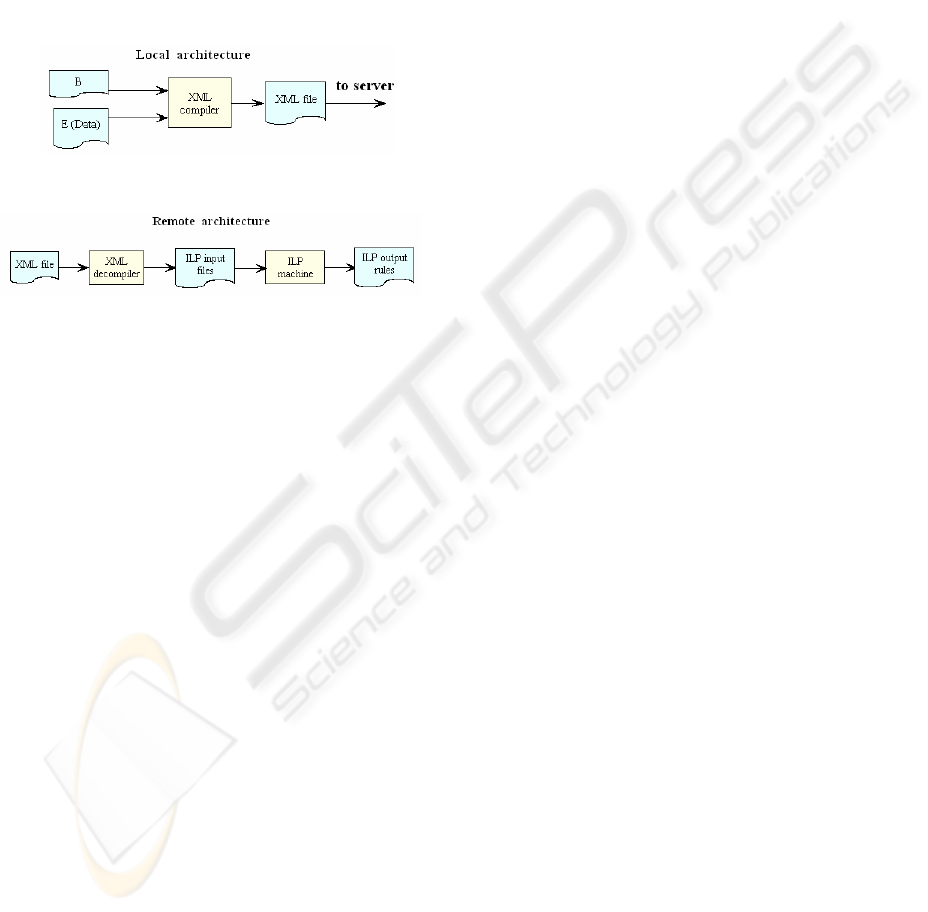
We have below two functional diagrams explaining
the flow of data and the expected output.
Figure 2
Figure 3
The output of the ILP machinery is a finite set of
logical rules that are translated into a human
readable format. The rules are output in an
implicative form and this is understandable as soon
as the predicate names support intuition.
5 CONCLUSION
Today, it is a common technique to enable a lot of
applications with e-technology. We investigate here
the field of Inductive Logic Programming targeting
to a biological application. Due to the similarity
between the first-order logic syntax and XML, there
is no difficulty to use XML as a standard format for
our files exchange. One of the main interest of our
approach is that we only manipulate data and files
which are almost human readable and the output
product (a set of rules) is easily understandable for
non-expert people. We expect to have a continuous
flow of data, and so to continuously improve the
target ILP machinery. As a mid-term objective, we
want to provide a public access to the machinery and
to make several ILP engines (see (Richard and
al.2004) for instance) to compete for extracting
rules. Since the underlying language is the same
(first order logic), it is very easy to compare and
why not, to mix different extracted rules to get a
better understanding of the process on hands. As a
long term objective, our platform could create a
vacuum of knowledge under one roof and open the
doors to a worldwide consortium of Bio Informatics
experts to access data, research, forums, resources
and application through the Internet.
REFERENCES
Farmer M., Zakariya M,.2004 : Enabling e-Technology for
bio-informatics. In: UK-Japan High Technology
Industry Forum, London.
Inoue, K. 2004, Induction as Consequence Finding. In:
Machine Learning, 55(2):109-135.
Inoue, K., Saito, H.2004: Circumscription Policies for
Induction. In: Proceedings of 14
th
Conf. on Inductive
Logic Programming, LNAI 3194, pp.164-179,
Springer.
Serrurier M, Prade D, Richard G.2004: A simulated
annealing framework for ILP. In: Proceedings of 14
th
Conf. on Inductive Logic Programming, LNAI 3194,
pp.288-304, Springer, September 2004.
Manyri L., Doncescu A., Benchaban F., Urribelarea J.L.
2005 : Parallel Differential Evolutionary Algorithms
for Parameters Estimation in Fermentation Process. In:
Journal of Parallel and Distributed Computing
Practice-SIAM
A WEB-BASED ARCHITECTURE FOR INDUCTIVE LOGIC PROGRAMMING IN BIOLOGY
361
