
INTEGRATION OF DATA SOURCES FOR PLANT GENOMICS
P. Larmande, C. Tranchant-Dubreuil, L. Regnier, I. Mougenot, T. Libourel
CIRAD, IRD, CINES, LIRMM, CNRS
UMR PIA, TA 40/03, av Agropolis Montpellier cedex 5, 34389, France
Keywords:
Plant genomics, interoperability, integration, mediator.
Abstract:
The study of the function of genes, or functional genomics, is today one of the most active disciplines in the life
sciences and requires effective integration and processing of related information. Today’s biologist has access
to bioinformatics resources to help him in his experimental research. In genomics, several tens of public data
sources can be of interest to him, each source contributing a part of the useful information. The difficulty lies
in the integration of this information, often semantically inconsistent or expressing differing viewpoints, and,
very often, only available in heterogenous formats. In this context, informatics has a role to play in the design
of systems that are flexible and adaptable to significant changes in biological data and formats. It is within
this framework that this paper presents the design and implementation of an integrated environment strongly
supported by knowledge-representation and problem-solving tools.
1 INTRODUCTION
The study of the function of genes, or functional ge-
nomics, is today one of the most active disciplines
in the life sciences. Researchers in the domain have
to manage a constant flow of voluminous information
originating from:
• experimental data,
• data that is structured or semi-structured according
to pre-existing adapted schemas (databases),
• analysis results,
• annotations,
• output from models, etc.
For example, research into mutations in an organism
is one of the study techniques of functional genomics.
To identify a mutation in a gene and the consequences
it can lead to, the biologist researcher has to consult
and compare several information sources such as bi-
ological sequencing data obtained from public or pri-
vate resources. He has also to compare experimen-
tal data and data that has undergone diverse proce-
dures to be able to refine and perfect his analyses.
Functional genomics is a research subject that there-
fore relies on several tools, expertise and resources to
process and produce information on the function of
genes. To this end, access to different necessary re-
sources and services is imperative. However, there
exist few integrating solutions for plant genomics and
this lack is as much felt here as it is within the bio-
medical community, whose situation is very similar
(Davidson et al., 1995).
One has to admit that the difficulties are many. To
identify, sort and annotate sources is a task that is be-
coming more and more difficult. From a biological
point of view, there is need (i) to identify the relevant
sources for an analysis from a catalogue that does not
stop growing in size (Galperin, 2004), (ii) to confer a
degree of confidence to a source and to possibly anno-
tate it, (iii) to keep track of regular updates and to pos-
sibly rerun one’s own analyses. From an informatics
point of view, one is faced with a diversity of formats
because, depending on the need and the concerned ex-
periments, the biological sources can have very het-
erogeneous structures. For example, data is often in
a flat file (e.g., Format GenBank, EMBL, ASN1) but
could also be in an XML file or in the form of tabu-
lated data. The structures of the sources also undergo
changes and can present variations.
Any attempt to integrate diverse data, procedures, ex-
pertise and resources leads to problems of syntactic
and semantic interoperabilities in the informatics do-
main. To these problems must be added the factor
that this information is often sensitive (even confiden-
tial) and highly variable and subject to change. The
goal of the present work is to present an integrated
environment for knowledge representation that per-
mits interoperability between different data sources
by overcoming problems of heterogeneity . The in-
314
Larmande P., Tranchant-Dubreuil C., Regnier L., Mougenot I. and Libourel T. (2006).
INTEGRATION OF DATA SOURCES FOR PLANT GENOMICS.
In Proceedings of the Eighth International Conference on Enterprise Information Systems - DISI, pages 314-318
DOI: 10.5220/0002455103140318
Copyright
c
SciTePress

tegration of data sources can be approached from dif-
ferent angles, each with its own advantages and draw-
backs (Karp, 2003); we present a brief state of the art
in the following section 2. Section 3 will be dedicated
to an operational mediation system: Le Select. In the
rest of the article, we will show how our exploratory
approach is based on the contextualization of this sys-
tem for use in the plant genomics domain.
2 STATE OF THE ART
The integration of heterogeneous data is a question
that has occupied the information-systems scientific
community for many years now. And the commu-
nity has responded in many ways with various solu-
tions. Within the confines of the database domain, the
approach has gone from the distributed perspective
to the federated databases perspective to the media-
tion perspective. The arrival of Web technologies has
also strongly influenced this concept of integration by
offering ’light’ integration solutions as well as more
complex solutions based on integrators of Web ser-
vices. Within the genomics context, the solution that
is the most handy is the light integration of sources
in their original formats. References cross-coded us-
ing hypertext links are now found in a large number
of biological sources (e.g., GenBank, SWISS-PROT)
and searching them by browsing allows fast access to
information.
Other, more complex, solutions integrate the sources
by offering a common interface and query language to
users. Several such systems have been implemented,
for example, SRS
1
, the Entrez
2
system developed at
NCBI, DBGet
3
in Japan or even the French ACNUC
4
system (Gouy et al., 1985). In all these examples,
the systems are analogous to huge access catalogues:
the user selects the sources he want to query from
amongst those already indexed.
Recently, proposals for stronger integration have been
put forward. Architectures that allow transparent ac-
cess to the user and the location of data sources by
integrating their schemas have been proposed. Sev-
eral solutions have emerged:
• non-materialized integration offered by mediation
architectures, most often designed around a single
representation model and a high-level query lan-
guage,
• ’materialized’ integration requiring the pre-
integration of the various data in data warehouses
whose schema is designed using the schemas of the
1
Sequence Retrieval System, www.infobiogen.fr/srs/
2
www.ncbi.nlm.nih.gov/Entrez/
3
www.genome.ad.jp/dbget/
4
pbil.univ-lyon1.fr/search/query.html
concerned sources and depending on the analyses
to be conducted.
In mediation systems, a mediator module manages ac-
cess to distributed sources. It breaks down the user’s
global query into elementary queries, assigning each
to a distributed source that can respond to it. It then
recomposes the response to the global query from re-
sponses to the elementary queries. Adaptors or ’wrap-
pers’ interpose themselves between each source and
the mediator. Systems developed according to this
architecture include K2/Kleisli, DiscoveryLink and
TAMBIS (Eckman et al., 2001). Access to biological
resources by these systems however remains limited
and does not fully satisfy scientists’ requirements.
On the other hand, in the materialized approach,
the data warehouses import locally the sources in
one same schema and the user’s global request is
processed directly. In France, in the plant genomics
domain, the bioinformatics platform G
´
enoplante-
info
5
uses this approach for integration (Samson et al.,
2003; D. Samson et al, 2005; A. Duclert et al, 2005).
This type of solution permits cleanup and annotations
of imported data (Susan B. Davidson et al, 2001).
Thanks to the rapid evolution of Web technologies,
applications can now develop Web-based services (re-
sponding, for example, to a specific query). These
Web services have the advantage of homogenizing the
data exchange format, thus facilitating interaction be-
tween applications. Moreover, they can be included
within a repository. This is the case with the Bio-
Moby repository project which implements a service
for locating resources based on a service ontology (P.
Lord et al, 2004). Another project, myGRID, helps
the user locate a sequence of services appropriate for
any one analysis.
In the following sections, we will describe the media-
tion system that we have used and the stages involved
in integrating the sources.
3 LE SELECT
Le Select is an integration system of type mediator
(Manolescu et al., 2002; Cavalcanti et al., 2002) de-
veloped at INRIA at Rocquencourt within the frame-
work of the Caravel project
6
. This system provides
uniform access for integrating, publishing and interro-
gating distributed heterogeneous sources by support-
ing several data types: structured and semi-structured
data, flat files, images, etc. It also handles programs
that use this data in the same manner. Le Select is a
mediator for access to distributed and heterogeneous
resources. A Le Select server authorizes the publica-
5
genoplante-info.infobiogen.fr
6
http://www-caravel.inria.fr/LeSelect/
INTEGRATION OF DATA SOURCES FOR PLANT GENOMICS
315
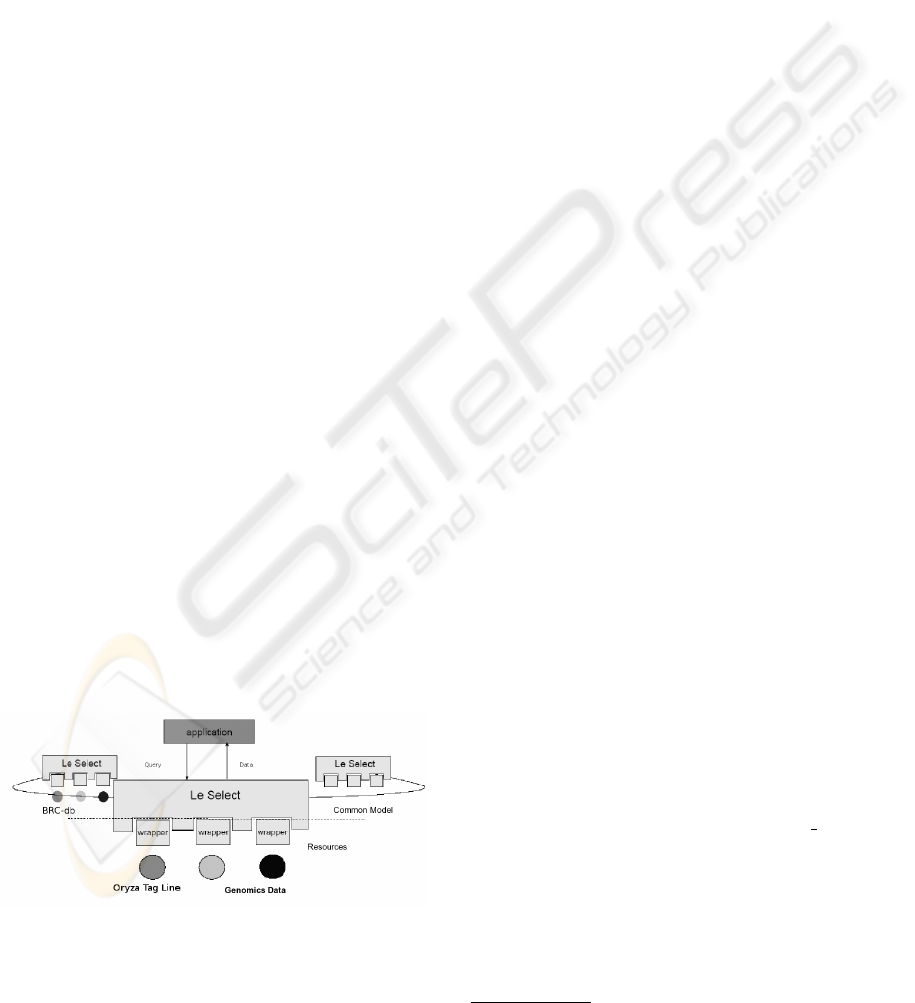
tion of a resource (data or program), while preserv-
ing the sources’ independence and format. It allows a
certain amount of flexibility of use since the resources
can be published incrementally in the system.
Before a resource can be published, an adaptor (wrap-
per) specific to the resource has to be designed (Fig.
1.). The creation of wrappers is the responsibility
of the administrator of the concerned resource but a
wrapper library dedicated to standard resource types
is slowly growing. Wrappers play multiple roles:
as translators between the mediator and the resource
by offering a homogenous representation based on
the relational model and as exporters of statistical
metadata on the resources they publish, such as the
resource’s availabilty, the query execution time, the
ability to execute a query (ability to join two resources
or to test them for equality).
As for the mediator, it offers a pivot language for
querying (close to the SQL standard). It includes a
query management and optimization program which
uses meta-information supplied by the wrappers to es-
tablish the execution plan of every query. At a general
level, it breaks down the global query into elementary
queries, assigning each elementary query to a source
that can respond to it, submits the queries, then recon-
structs the complete response from the elementary re-
sponses.
Finally, since the Le Select server is installed on a
Web server, it possesses an interface which can be ac-
cessed by a Web browser. Published resources are
thus accessible by simple browsing (access by links)
or by the intermediary of SQL queries.
The fact that Le Select communicates using SQL is
of great interest because existing applications can be
reused to connect to sources. For example, an applica-
tion can be connected to the server network and com-
municate with a mediator via a JDBC bridge (Fig. 1).
At the same time, several servers can be installed
within the network (specially P2P peer-to-peer net-
works) and can co-operate to provide access to data
and services.
Figure 1: Diversity of resources that can be published.
3.1 Contextualization of Le Select
In our context, the exchange and distribution of
information for the purposes of sharing it is of the
utmost importance. Not only does data exchange
allow the validation of data by other scientists
running identical analyses, but data sharing also leads
to generation of new data. Within the framework of a
functional genomics project whose goal is to generate
a collection of rice insertion lines (Oryza sativa)
(Sallaud et al., 2004), a database has been created to
store experimental data: Oryza Tag Line (OTL)
7
.In
addition, another database, Rice BRC-db (BRC-db),
has been developed for consolidating information on
genetic and genomic rice resources.
The short term objective is to bring closer together
the correlated information in these two databases,
indeed to migrate part of the information from OTL
to BRC-db, within the strict condition that the two
databases should remain independent (DBMS and
interface). In such a scenario, using a mediator
approach seemed of interest to us.
Before publishing the two information systems,
we designed a virtual global conceptual model to
solve the problems of semantic heterogeneity and
to define correspondences between the entities. We
encountered, for example, a case of homonymy
(Karp, 1995): the class ’plant’. In the OTL model,
this referred to ’mutant’ plants whereas in BRC-db
they were ’wild’. In this case, a new ’plant’ class
was created in the global schema to correspond to
the ’mutant’ individuals. To help limit problems of
this nature, we referred to shared definitions within
the domain: the ontologies (Gruber, 1993). The
ones that are most commonly used in plant genomics
are Gene Ontology (M.A. Harris et al, 2004) and
Plant Ontology (The Plant OntologyTM Consortium,
2002).
Figure 2 shows the publication of OTL via Le Se-
lect’s interface. As can be seen, the page is divided
into four parts. On the left are displayed the wrappers
corresponding to the published sources. At the top,
one can select the types of wrappers (wrappers & ta-
bles, views and programs). In the central part of the
screen is displayed the data corresponding to a query
posed in the Query area at the bottom. In the example
shown, data is displayed from the TRAIT
VIEW ta-
ble corresponding to the phenotypical characters ob-
served in the collection of mutants. Data is directly
extracted from the database, the table structure is not
modified. The wrapper created for publishing this re-
source uses Java drivers (JDBC).
As we wanted to bring together data from the two
databases in conformance with the studied virtual
7
http://genoplante-info.infobiogen.fr/OryzaTagLine
ICEIS 2006 - DATABASES AND INFORMATION SYSTEMS INTEGRATION
316

Figure 2: Oryza Tag Line published by Le Select.
global conceptual model, we would have had to trans-
form the schema of the sources before publication.
But Le Select also offers a mechanism for viewing
published sources. For the mediator, the views are
also wrappers that execute a query on an already-
published source. It is this feature that we have used
by creating views of tables that have to be trans-
formed. This establishes, in a simple manner, the cor-
respondences between the OTL and BRC-db database
schemas.
4 CONCLUSION
The information systems that researchers in func-
tional genomics have to put together to fulfil their re-
quirements need to preserve the resources’ indepen-
dence and, very often, the confidentiality of at least a
part of their information.
The mediation solution is thus of relevance; it con-
serves the resources’ independence while allowing
their distribution and it provides uniform access to in-
formation. In fact, even though the materialized ap-
proach is also a robust one, it does not handle well the
changing character of genomic sources. Unavoidable
changes in both systems, OTL and BRC-db, would
entail numerous changes to the schema of the data
warehouse and, subsequently, to the procedures for
loading the underlying data.
The solution implemented using Le Select takes
changes in the Oryza Tag Line schema in stride; they
are incorporated directly by the mediator. And, by
using the intermediary of views, the establishment of
new correspondences with BRC-db is also relatively
easy. We can thus think that the approach we propose
is transferable to other functional genomics applica-
tions. On the longer term, aside from incorporating
the integration of programs, the systems should allow
researchers to conduct online analyses by authorizing
procedures on the data (access to both types of re-
sources having been made transparent).
REFERENCES
A. Duclert et al (2005). Bioinformatics in Genoplante.
Plant Genomics European Meetings proceedings.
Cavalcanti, M. C., Mattoso, M., Campos, M. L., Llirbat, F.,
and Simon, E. (2002). Sharing scientific models in
environmental applications. In SAC ’02: Proceedings
of the 2002 ACM symposium on Applied computing,
pages 453–457, New York, NY, USA. ACM Press.
D. Samson et al (2005). GpiIS: Towards an integrated infor-
mation system around plant genomes. Plant Genomics
European Meetings proceedings.
Davidson, S., Overton, C., and Buneman, P. (1995). Chal-
lenges in integrating biological data sources. J Com-
put Biol, 2(4):557–72.
Eckman, B., Lacroix, Z., and Raschid, L. (2001). Optimized
seamless integration of biomolecular data. IEEE sym-
posium on Bio-Informatics and Biomedical Engineer-
ing (BIBE’01), Washington DC, pages 23–32.
Galperin, M. (2004). The Molecular Biology Database Col-
lection: 2004 update. Nucleic Acids Res, 32(Database
issue):D3–22.
Gouy, M., Gautier, C., Attimonelli, M., Lanave, C., and
di Paola, G. (1985). ACNUC–a portable retrieval
system for nucleic acid sequence databases: logical
and physical designs and usage. Comput Appl Biosci,
1(3):167–72.
Gruber, T. (1993). Towards principles for the design of on-
tologies used for sharing. The International Workshop
on Formal Ontology.
Karp, P. (1995). A strategy for database interoperation. J
Comput Biol, 2(4):573–86.
Karp, P. (2003). What database management system(s)
should be employed in bioinformatics applications?
OMICS, 7(1):35–6.
M.A. Harris et al (2004). The Gene Ontology (GO) data-
base and informatics resource. Nucleic Acids Res,
32(Database issue):D258–61.
Manolescu, I., Bouganim, L., Fabret, F., and Simon, E. (Jan
2002). Efficient querying of distributed resources in
mediator systems. In Lecture Notes in Computer Sci-
ence, volume 2519, pages 468 – 485.
P. Lord et al (2004). Applying semantic web services
to bioinformatics experiences gained, lessons learnt.
ISWC Springer-Verlag Berlin Heidelberg, pages 350–
364.
Sallaud, C., Gay, C., Larmande, P., Bes, M., Piffanelli, P.,
Piegu, B., Droc, G., Regad, F., Bourgeois, E., Mey-
nard, D., Perin, C., Sabau, X., Ghesquiere, A., Glasz-
mann, J., Delseny, M., and Guiderdoni, E. (2004).
High throughput T-DNA insertion mutagenesis in
INTEGRATION OF DATA SOURCES FOR PLANT GENOMICS
317

rice: a first step towards in silico reverse genetics.
Plant J, 39(3):450–64.
Samson, D., Legeai, F., Karsenty, E., Reboux, S., Veyri-
eras, J., Just, J., and Barillot, E. (2003). Genoplante-
info (GPI): a collection of databases and bioinformat-
ics resources for plant genomics. Nucleic Acids Res,
31(1):179–82.
Susan B. Davidson et al (2001). K2Kleisli and GUS: Exper-
iments in integrated access to genomic data sources.
IBM Systems Journal, 40(2):512–31.
The Plant OntologyTM Consortium (2002). The Plant On-
tologyTM Consortium and Plant Ontologies. Com-
parative and Functional Genomics, 3(2):13.
ICEIS 2006 - DATABASES AND INFORMATION SYSTEMS INTEGRATION
318
