
GENERATION AND USE OF ONE ONTOLOGY FOR
INTELLIGENT INFORMATION RETRIEVAL FROM
ELECTRONIC RECORD HISTORIES
Miguel A. Prados de Reyes, M. Carmen Peña Yañez, M. Amparo Vila Miranda,
M. Belen Prados Suarez
Computer Science and A.I. Department. University of Granada,Computer Science Department. San Cecilio Hospital,
Granada,Computer Science and A.I. Department. University of Granada,Computer Science Department.University of Jaen
Keywords: Information Retrieval System, Semantic Information, Semantic Organization, Ontology, Electronic Health-
care Record.
Abstract: This paper analyzes the terminology used in the diagnosis, treatment, exploration, and operation descrip-
tions entered by doctors in the electronic healthcare record. From this, expression stability (and the use of a
sufficiently limited and controlled language) is shown, which is therefore reasonably valid for a conceptu-
alization process to be employed on it. This conceptualization process is performed by the generation of an
ontology which proposes semantic classes according to the different medical concepts to be used on data-
base query profiles. By way of summary, we shall propose a semantic organizational method so that
classes, attributes and properties in the ontology may act as links between the database and the users, both
in information incorporation processes and in queries. It offers a wide range of benefits by extending and
making information management possibilities more flexible, and enabling the application of traditional data
mining techniques.
1 INTRODUCTION, STARTING
HYPOTESYS
Existing Medical Information Retrieval Systems
(MIRS) present certain problems and drawbacks as
they usually require deterministic query profiles
which use structured data sets that do not always
meet the real requirements of the staff accessing the
medical information database (Prados, 2003). This is
why documentary searches are not very thorough
and have a lot of noise (Noy, 2003). In the majority
of cases, the most interesting medical information is
to be found in the doctor's own notes, but queries on
these are not always successful as the information is
unstructured and there are the usual problems of
natural language and its formalization.
An observation of the expressions describing di-
agnoses, treatments, interventions and explorations
reveals the following characteristics: They are short;
They contain the most relevant medical aspects;
They are repetitive and have a limited language.
These characteristics suggest that it is possible to
analyze the terminology used, the composition and
the structure of these phrases in greater depth,
thereby demonstrating that: its terminological do-
main is limited; the conceptual domain is suffi-
ciently strict and categorized; phrase composition
and structure is stable.
This leads us to approach the generation of an
ontology by establishing a conceptualization based
on an in-depth analysis of the language and the
terms used, and this can be used in the data capture
and information retrieval process. The ontology
must be constructed in such a way that it can charac-
terize the diverse semantic contents that an expres-
sion entails.
2 BACKGROUND
The background deals with two closely related top-
ics. Traditional "documentary techniques" generate
an entire set of techniques and tools in their digital
management which is the object of documentary
computer science (Maniez, 1992).
Nowadays, there is a wider opinion about the
problem which is based on so-called "Text Mining"
(TM), specially motivated by interest in the "seman-
565
A. Prados de Reyes M., Carmen Peña Yañez M., Amparo Vila Miranda M. and Belen Prados Suarez M. (2006).
GENERATION AND USE OF ONE ONTOLOGY FOR INTELLIGENT INFORMATION RETRIEVAL FROM ELECTRONIC RECORD HISTORIES.
In Proceedings of the Eighth International Conference on Enterprise Information Systems - ISAS, pages 565-571
DOI: 10.5220/0002465205650571
Copyright
c
SciTePress

tic web". Like the data mining definition, text min-
ing may be defined as: "The entire process of ex-
tracting relevant information that is not explicitly
present in a document collection". There is a clear
distinction between this and documentary computer
science, which only attempts to show explicitly
present concepts (Delgado, 2002).
In text mining, it should be remembered that we
are dealing with information which is not particu-
larly structured, and therefore traditional data min-
ing techniques cannot be applied. This lack of struc-
ture is the biggest problem for TMs and requires the
texts to be preprocessed and converted into an "in-
termediate form" so that the algorithms and methods
(classification, association, etc.) being used may be
applied to them.
Current study of ontologies and their design and
development tools (Lambrix,2003) has produced
new methods and techniques, but mainly a wider,
more ambitious vision in textual data processing,
since they make action possible on previously con-
ceptualized domains. This is particularly interesting
as here there is an intermediate element that can be
used as a link between the "gross" textual informa-
tion and its preprocessing and subsequent treatment
using usual data mining techniques. More specifi-
cally, the ontology is particularly important in in-
formation retrieval systems since they provide the
connection between final user applications and data-
bases and, contrariwise, in visualization processes
(Guarino,1998 ).
In short, it is clear that this is an interdisciplinary
field which includes elements, methods and tech-
niques from documentary computer science, linguis-
tics, and data mining, with their contributions relat-
ing to information retrieval, information extraction,
clustering, categorization and automatic learning
(among others) (Prados,2004).
3 WORK DESCRIPTION
The basic idea behind this work is that the medical
language used in diagnostic expressions and deter-
minations (based on natural language) is sufficiently
controlled and strict with an adequately formal
grammar as to be worth using in data processing and
documentary searches which could be applied di-
rectly on the medical language without the need for
code systems. This would make documentary
searches more powerful by enabling descriptive and
qualifying aspects to be included in the pathology or
treatment, and not merely unqualified searches on
the diagnosis (Shankar, 2002). A diagnostic expres-
sion comprises terms with a semantic content which
are relevant for diagnosis. It is first necessary to
analyze the semantic typology of these terms and to
establishing the different classes to which a lexical
unit can belong.
Our aim in this paper is to show that this is pos-
sible. In order to do so, we have considered four
classes of term sets: identifiers, locators, qualifiers,
and etiologicals. From this first semantic structure,
we can extract an ontology which conceptually
describes the domain of the medical diagnosis, and
propose a methodology for information storage and
retrieval, extending the restrictive possibilities of
traditional searches on the electronic healthcare
record using the defined ontology.
Our first objective is to define what we call the
"semantic classes", each of which represents a kind
of concept used by doctors in their descriptions. Our
second objective is to check that the medical expres-
sions in natural language are in keeping with the
element coordination of each semantic class. Our
third objective is to confirm the hypothesis that the
constituent elements of each semantic class repre-
sent a perfectly determinable and controllable set. A
class may have superclasses which offer a medical
meaning membership from one given concept to
another with a more general meaning (Dameron,
2004).
We also define what we call the "semantic class
sequence" (SCS) as the sequence obtained in the
identification of the semantic classes from an ex-
pression, and their sequence of presentation. By way
of example, the SCS “I-E-L” means "identifier-
etiological-locator". Although the purpose of these
SCSs is to check stability in expression composition,
they will also be useful in both the data acquisition
process and the documentary search process from
the data structures of the semantic classes.
The system is enriched by extracting the corre-
sponding ontology (ONTOARCHINET), its hierar-
chies and properties, with it having a welldefined
conceptual environment and making its use avail-
able for the documentary search.
We shall also discuss whether this proposal is
worthwhile and if it is possible to construct a set of
data structures representing the members of each
semantic class and their properties (associations,
frequency, medical environment, etc.) so that under
the analyzed (cleaned and filtered) documentary
search, the SCS can be recognized. Its components
are then extracted in order to perform the search,
focusing or extending it according to the semantic
components in the ontology.
ICEIS 2006 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
566

4 PREVIOUS MEDICAL
TERMINOLOGY RESULTS:
EXAMPLE OF CARDIOLOGY
A search was performed on our electronic healthcare
record system (relating to admissions between 2000
and 2004) for medical records listing cardiology in
their documentary profile. The number total of ex-
pressions found in the period of time is, 433.492;
Related to cardiology 39.781; and syntactically
different 1.656.
Two expressions are considered syntactically
different when there is some variability in the terms
they comprise or in their order. At this point, we
should introduce the concept of "medically different
expressions". By syntactically grouping different
expressions according to this criteria, we obtain 627
medically different expressions. On examination of
the results obtained, we find that the number of high
frequency expressions is greatly reduced, and that
the frequencies of the others are less relevant. This
suggests that the language is unified to a large ex-
tent.
In order to provide our system with a value rep-
resenting the differentiation degree of expressions,
we define an index (which we call the “diversifica-
tion index”) as the quotient between the number of
syntactically different expressions and the number
of different medical expressions. By analyzing the
results, we can extract the following fundamental
considerations:
The number of medically different expressions
does not obviously differ from the number of syn-
tactically different expressions. This confirms the
previously indicated language unification.
As the number of expressions increases, diversi-
fication tends to decrease or to stabilize. This sug-
gests that the system is stable: if we increase the size
of the study period, the obtained results regarding
differentiation would not be significantly different
from the ones presented here.
In the second stage, the semantic composition of
the expressions is analyzed by studying the terms
comprising them. This analysis suggests the exis-
tence of a small number of the following specific
semantic categories:
Identifiers: Description terms with maximum
rank within the expression, absence of which deter-
mines its lack of logical sense. Such a descriptor
necessarily refers to a pathological process, opera-
tion or exploration.
Qualifiers: Secondary terms that specify the
meaning of the identifier. On their own, they have
no clinical meaning but they indicate a medical
qualification: “serious", "acute", "spontaneous", etc.
These qualifiers can, in turn, have different typolo-
gies such as those qualifying severity, time, com-
pensation or location.
Etiologicals: Terms representing the origin or
causes of the pathological process (when it exists).
Some examples we can mention are accidental poi-
soning, infections or age, and these appear in the
main diagnosis as the cause.
Locators: These represent the anatomical place
of the human organism where the damage or injury
appears and the identifier takes place (such as auri-
cle, ventricle, or myocardium, etc. for the specialty
that this work focuses on).
The number of descriptors found for each cate-
gory is: 115 for identifiers, 167 for qualifiers, 29 for
etiologiclas, and 95 for locators.
The vocabulary is clearly small enough for data
management not to be excessively difficult. Al-
though we have considered the possibility of ignor-
ing very low frequency descriptors, we do not be-
lieve this to be appropriate since there are patholo-
gies with a very low frequency but with a very high
medical interest. In addition, a kernel of high fre-
quency members can be seen, with a considerably
different frequency regarding the classes outside this
kernel. The border comprises a much reduced num-
ber of members.
5 “ONTOARCHINET”
ONTOLOGY EXTRACTION
According to the terminological study undertaken, it
is necessary to establish which the classes of the
ontology and their possible hierarchizations are in
accordance with the generality character. For this
purpose, utility character must prevail in order to
offer access to the information depending on the
elements in each class and their possible priorities.
The ontology focuses on documentary search using
the concepts contained in the free-text diagnosis
descriptions used by doctors.
The stages of the ontology construction process
are: Determination of domain and scope; Definition
of domain concepts (classes); Hierarchical classifi-
cation of concepts (subclasses–superclasses hierar-
chy) Definition of class properties (slots) and their
value restrictions;Definition of instances, according
to the defined properties.
5.1 Scope and Domain
Determination
As seen before, the domain is limited to diagnosis
expressions: summary representations relating to a
diagnosis, treatment, operation or exploration de-
GENERATION AND USE OF ONE ONTOLOGY FOR INTELLIGENT INFORMATION RETRIEVAL FROM
ELECTRONIC RECORD HISTORIES
567
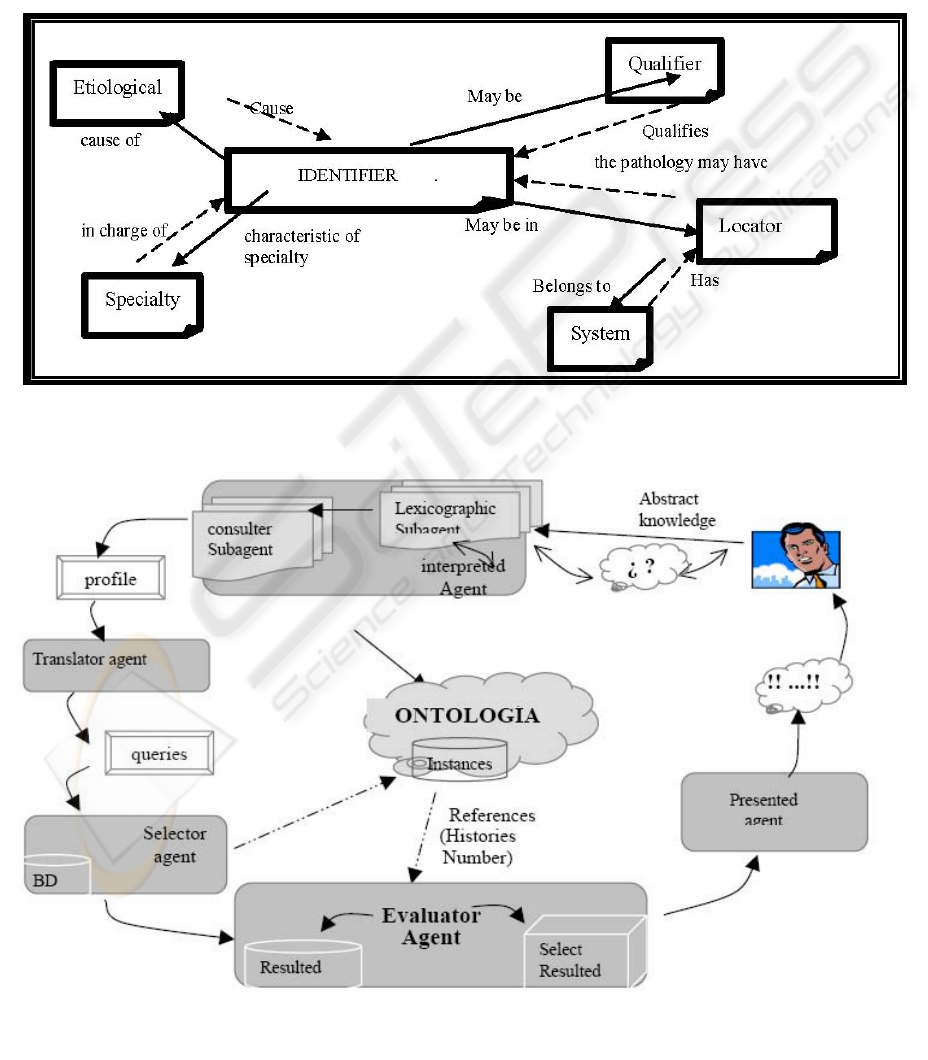
termined by doctors involved in a health care proc-
ess (consultation, clinical admission, urgent assis-
tance, etc.). There may be several expressions within
any process. The domain of this ontology reaches
expressions such as "aborted myocardial infarction"
or "previous acute extensive myocardial infarction".
Several queries may be done in this field like:
• Could we determine the characteristics for a
given identifier?
• Is a given descriptor cause or effect?
• Which patients have suffered an identifier
for a severity qualifier?
• Does a given semantic class have the etio-
logical as a secondary associated cause?
The role of the ontology is two-fold: firstly, it is
a place to store expressions which it has classified
terminologically and semantically, according to the
established classes and hierarchies; and secondly, it
enables documentary search in accordance with the
established semantic classes and their corresponding
hierarchies and properties.
Figure 2: General query scheme.
Figure 1: Relationship general scheme.
ICEIS 2006 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
568

5.2 Concept Definition in the
Domain (classes)
These concepts must be those used in diagnosis
expressions and also those delimiting the scope of
the medical specialty. In addition, the concept of
descriptor type according to the established catego-
ries mentioned in the previous section must also be
introduced.
It is clearly of clinical interest to consider essen-
tial factors describing a pathology which is accom-
panied by relevant descriptions. In this regard, the
categories established for terminological study seem
appropriate: identifiers, qualifiers, etiologicals and
locators (Figure 1).
The target of these main classes is to define se-
mantic concepts. However, it is possible to establish
other classes which are useful for the documentary
search and acquisition process in addition to others
which are directed towards the structuring of differ-
ent medical fields such as: Medical specialty , Sys-
tem (human ana-tomy) and Infraconcepts (some
terms that become representative when others are
present.
5.3 Hierarchical Classification of
Concepts
Although, in principle, hierarchies are established
according to the generality levels of each descrip-
tor’s semantic concept, the generalist nature we may
encounter might require character “subtype” sub-
classes to be defined.
For each class, we have established the levels we
consider strictly necessary, thereby avoiding their
proliferation which could make ontology manage-
ment uncomfortable. In the following sections, we
shall present the hierarchization for each of the main
classes considered.
Identifiers: In this case, the hierarchy is strictly
set acording to a sense of generalitation or speciali-
zation from a strictly clinical point of view.
Qualifiers: This class suggests the existence of
“subtype” members: qualifier classes established
according to the character they qualify. We can then
find qualifiers such as: Severity: qualifies how the
pathology affects the patient; Relevance: qualifies
the degree to which the pathology is present; Com-
pensation: stability in development and presentation;
Clinical: qualifies the existence of associated pa-
thologies such as “obstructive ischemia” ; Timing:
qualifies the chronicity level.
From this first level including different qualifier
sub-types, hierarchies are established according to
the level of generality.
Etiologicals.- Similarly, we should examine the
different types of causes producing the illness. We
should consider three initial subtypes: external
causes, secondary (to other pathologies) causes, and
internal causes (metabolic, physiologic). In order to
avoid an excessive number of levels in the ontology,
internal causes have been included in the secondary
causes. External causes have a wide typology de-
pending on whether they refer to habits, accidents,
external agents, etc.
Locator: The purpose of this class is to define
the affected anatomical area; the anatomical areas
must then be classified. It is then necessary to decide
which of the commonly used hierarchization criteria
to follow. From the point of view of the ontology,
this is a secondary topic since what we want to iden-
tify is an area rather than its function (this might be
a property in the ontology).
Infraconcepts: Because of the usefulness and
ease of use of the ontology, we consider this class in
order to clarify the meaning of the concepts repre-
sented by other classes in the ontology. Initially, we
only considered one kind of infraconcept (the so
called infralocator) that enables us to precisely de-
termine an anatomical location (e.g. the “internal
side” of a given anatomical locator). The infraloca-
tor name is given by the ambiguous meaning, and
needs to be joined to a locator class member in order
to have a specific meaning, such as in “ventricle
internal side”.
The existence of more infraconcepts is an open
topic in the ontology, since we have only considered
the Infralocator infraconcept.
Once concepts have been defined according to
the class structure outlined above, we propose an
implementation in "Protégé", the tool used to define
our ontology.
5.4 Definition of Class Attributes
and Properties (slots), and
Restrictions on Their Values
The properties (intrinsic properties) represent value
relationships with other classes (Figure 1). There
may also be extrinsic properties which provide
added values for the corresponding instances which
are meaningful from the clinical or medical man-
agement point of view.
Looking at the intrinsic properties, the slots in
the class definition describe the class instance attrib-
utes and relationships with other instances. By way
of example, each identifier is then related to a cause
and anatomical area. An anatomical location may
also be linked to a certain qualification. The purpose
of these slots is to establish relationships and restric-
tions between the different class instances. This
GENERATION AND USE OF ONE ONTOLOGY FOR INTELLIGENT INFORMATION RETRIEVAL FROM
ELECTRONIC RECORD HISTORIES
569

enables class hierarchy navigation in the documen-
tary search but assuming restrictions which give
coherence to the possible profiles designed.
Designing these properties and relationships is
laborious and complex since they refer to objects
from other classes. So far, we have been referring to
intrinsic or extrinsic properties.
5.5 Instance Definition
Instances are each of the terms extracted during
previous linguistic analysis once the class they be-
long to has been typified. As mentioned previously,
the set is small enough for it to be managed easily.
Class-subclass hierarchization facilitates this by
reducing the number of direct final instances ob-
tained.
Two problems arise when these instances are de-
fined. The first problem is the need for medical
knowledge in order to define an instance member-
ship to a given class-subclass, particularly in the
identifier class. This is important since a wrong
definition would imply loss of exhaustiveness in the
documentary search. In such a case, the only solu-
tion is technical criteria. According to the method
we have developed, however, instantiation comes
from the very terminology and phrases used by
doctors and their classification according to ICD-9-
OMS. All of this reduces the error possibilities to a
minimum, and these are no greater than the differ-
ences that we would find if two specialists were to
construct the class-subclass instance structure. The
second problem is the treatment of documentary
synonymy. In order to solve this problem, before
defining the ontology, a previous and exhaustive
lexical analysis is needed (as we have done),
whereby this synonym is established using medical
supervision criteria.
6 USAGE METHODOLOGY
The use of the ONTOARCHINET ontology has
three levels of interest: acquisition process and data
storage; documentary query; moving through the
ontology
6.1 Acquisition Process and Data
Storage
Using the expression in natural languages, once it
has been filtered and cleaned, the terms with seman-
tic meaning can be matched to concepts in the on-
tology. In this process, the expression is expressed
using the concepts in the ontology and adding the
additional information stored in the ontology.
In the matching process, we also obtain a se-
quence of semantic SCS classes. These can be stored
in the database and used to calculate concept rela-
tion frequencies and the depth of each concept in the
ontology. By way of example, let us suppose the
expression in natural language: "Congestive cardiac
insufficiency due to acute lung edema". We would
obtain two expressions from this in the lexico-
graphic analysis: "Congestive cardiac insufficiency
due to acute lung edema" and “acute lung edema",
which would be described by means of the classes:
I=Insufficiency, L=Heart, C=Congestive, E=Edema,
C=Acute, L=Lung with SCS = ICLECL; and
I=Edema, C=Acute, L=Lung, with SCS=ICL, for the
second expression. Both the structured data and the
expression in natural language would be stored in
the database.
6.2 Documentary Query
The documentary query acquires the capacity of
multiple flexible queries which are very useful for
users. The process attempts to establish the concepts
contained in a query profile. Once these concepts
have been determined, selection on the database will
be made according to the profiles generated using
the hierarchical structure of classes and properties in
the ontology. This provides the query process with
flexibility and exhaustiveness and also frees it of
noise. The general scheme of user access is shown
in Figure 2, together with the different agents in-
volved in the query. Rather than exploring the read-
dressing possibilities in the query, or profiles of
forced sequences, exact or generic searches, we
shall simply say that all of them are perfectly feasi-
ble.
6.3 Moving through the Ontology
The possibility of moving through the structure of
the ontology is the first objective of the link with the
database, but it can also contribute to knowledge
about the relations and properties contained in the
ontology, which represents a way to access the data
structure and therefore build profiles by selecting
members and properties from the ontology. Depend-
ing on the extrinsic properties included, the ontology
will be able to answer a certain type of query, and
not only act as a link with the database. The great
advantage of the proposed method is that it does not
require a totally built or static initial ontology, and
therefore it can be improved as new expressions,
terms, concepts and relations arise, and this requires
ICEIS 2006 - INFORMATION SYSTEMS ANALYSIS AND SPECIFICATION
570

there to be an interface for the user specializing in
the maintenance work.
7 CONCLUSIONS
Our main objective was to establish the foundations
for developments that would allow information to be
processed using natural language. In our opinion,
this could then be used for terminology in the area
of cardiology:
• The structured sample is representative of the
system since the diversification index tends to
become stabilized as the number of cases in-
creases, and diversification is not excessively
high for the heads of series studied.
• By classifying the terms, the size of the classes
allows comfortable indexation of the terms.
Structures of classes-subclasses can be gener-
ated
• The physical data model can easily be generated
in the computer system by means of the corre-
sponding class tables.
• The classes and instances are enriched by means
of properties offering a greater richness of se-
mantic interpretation and more possibilities to
focus on the queries or extend them.
• The classes are sufficiently short and controlla-
ble, thereby ensuring process execution effi-
ciency
• The system allows an appreciably more powerful
documentary search than the traditional access
system using ICD-OMS codes. The tedious
process of codifying each expression is not nec-
essary
• Redundant fields in the tables are not needed to
codify contents which are already in expres-
sions provided by the doctor.
• Class and member properties can be added so that
the search process can be enriched by howing
these properties or using then in decision proc-
esses.
• The system can be enhanced by the incorporation
of new terms and expressions. In order to con-
sider more classes than those presented in this
paper, the semantic richness of the system
would be increased.
REFERENCES
Dameron O., Gibaud B., & Musen M. A. 2004. Using
semantic dependencies for consistency management of
an ontology of brain-cortex anatomy. First Interna-
tional Workshop on Formal Biomedical Knowledge
Representation KRMED04, Whistler, Canada.
Delgado M., Martin-Bautista M.J., Serrano J.M., Sanchez
D., Vila M.A., 2002. Association Rules extraction for
Text Minino. Lectures Notes in Artificial Intelligence
V. 2522 pp.154-162
Guarino N., 1998. Formal Ontology and Information
Systems. Formal Ontology in Information Systems.
pp. 3-15. (FOIS 98). Trento (Italia). IOS Press Weav-
ing the Biomedical Semantic Web with the Protégé
OWL Plugin. First International Workshop on Formal
Biomedical Knowledge Representation, Whistler,
Canada.
Lambrix P., Habbouche M., Perez M., 2003. Evaluation of
ontology development tools for bioinformatics.
Bioinformatic Vol. 19, n.12 pag 1564-1571.
Maniez J., 1992. Los lenguajes documentales y de
indización. Fundación Germán Sánchez Ruperez.
Pirámide, Madrid.
Martin-Bautista M.J., Sanchez D, Chamorro-Martinez J.,
Serrano J.M., Vila M.A. 2004. Mining Web Docu-
ments To Find Additional Query Terms Using Fuzzy
Association Rules. Fuzzy Sets and Systems V.148 pp.
85-1104.
Noy N., Tu S., 2003. Developing Medical Informatics
Ontologies Using Protégé Tutorial materials for AMIA
2003 tutorial.
Prados M., Peña M.C. 2003, Sistemas de Información
hospitalarios. Organización y gestión de Proyectos.
EASP (Escuela Andaluza de Salud Pública), Granada,
1
nd
edition.
Prados M., Peña M.C., Prados M.B., Garrido J.M. 2004,
Gestión de conocimiento en el ámbito hospitalario.
EASP (Escuela Andaluza de Salud Pública), Granada,
1
nd
edition.
Shankar R. D., Tu S. W., & Musen M. A.,2002. Use of
Protégé-2000 to Encode Clinical Guidelines. SMI-
2002-0944.(http://protégé.stanford.edu/doc)
GENERATION AND USE OF ONE ONTOLOGY FOR INTELLIGENT INFORMATION RETRIEVAL FROM
ELECTRONIC RECORD HISTORIES
571
