
AN ONTOLOGY-BASED INFORMATION SYSTEM FOR
MULTICENTER EPIDEMIOLOGIC STUDIES ON CANCER
J. M. Vázquez
1
, M. Martínez
1
, M. G. López
1
, B. González-Conde
2
, F. M. Arnal
2
J. Pereira
1
and A. Pazos
1
1
Medical Informatics and Radiological Diagnosis Center (IMEDIR), University of A Coruña, A Coruña, Spain
2
Universitary Hospital Complex Juan Canalejo (CHUJC), A Coruña, Spain
Keywords: Cancer, information systems, ontologies, epidemiology, handheld devices, security.
Abstract: Diseases like cancer are caused by a diversity of different factors interacting together, whose study requires
a huge amount of data. Compiling this data is an expensive and time-consuming task that can be carried out
in an easier, faster and more secure way with the support of Information and Communication Technologies
(ICT). Nevertheless, the majority of epidemiologic studies are executed without this support of informatics
or with basic tools that are developed by unqualified professionals. As a consequence, the integrity of the
collected data cannot be assured, and the reliability of the studies is usually decreased. This work presents
an ontology-based Information System for the development of multicenter epidemiologic studies on cancer
that allows 1) collecting, storing and editing medical data from different hospitals and 2) reusing the
compiled data by means of their integration with data from other systems. This system has been
satisfactorily applied to an epidemiologic study of colorectal cancer in Galicia, Spain.
1 INTRODUCTION
Epidemiologic studies are useful to understand the
origin of diseases, to detect outbreaks of pathologies
in the population, to make decisions to optimize
resources and, in sum, to improve the welfare and
quality of life for societies worldwide. However, the
study of multifactorial diseases like cancer, which
are caused by a variety of genetic, environmental
and lifestyle factors requires a large amount of data.
Compiling these data is a laborious work that
implies: a) the recovery of the patient’s clinical
records, b) the recovery of analysis data, and c)
personal interviews with the patient and his family
in order to fill out various questionnaires (e.g. family
questionnaires, risk factor questionnaires, etc.). In
addition, medical personnel in hospitals is usually
under a lot of attendance pressure and it is very
difficult for them to devote time and energy to
arduous tasks of interviews and information
collection. Therefore, facilitating as far as possible
this work becomes a critical task.
Information and Communication Technologies
(ICT) allow the development of Information
Systems (ISs) that provide mechanisms for the data
collection (including validation rules and error
control), as well as the storage and editing of
medical data from various hospitals by means of the
Internet. ICT allow us to efficiently manage large
amounts of data and therefore enhance the quality of
epidemiologic studies. It is also possible to use ICT
and the Internet to integrate various data sources and
as such obtain an even larger set of data.
In spite of these obvious advances, there still
exists a problem that concerns a great part of
epidemiologic studies: they are usually either carried
out manually, or using basic tools that have been
developed by unqualified personnel (it is only during
the data analysis phase when professional tools tend
to be used). Moreover, these tools are rarely
designed to allow their integration with other
systems. In consequence, the integrity of the
collected data cannot be guaranteed and the
reliability of the studies is frequently decreased.
133
M. V
´
azquez J., Mart
´
ınez M., G. L
´
opez M., Gonz
´
alez-Conde B., M. Arnal F., Pereira J. and Pazos A. (2008).
AN ONTOLOGY-BASED INFORMATION SYSTEM FOR MULTICENTER EPIDEMIOLOGIC STUDIES ON CANCER.
In Proceedings of the First International Conference on Health Informatics, pages 133-139
Copyright
c
SciTePress
2 BACKGROUND
The region of Galicia, situated in the northwest of
Spain, represents an excellent geographical area for
carrying out genetic-epidemiologic studies of colon
and rectal cancer due to the homogeneity of its
population in several dimensions: genetically (which
facilitates this type of studies) as well as culturally
and environmentally (which allows for conducting
homogeneous recruiting subjects that would
participate in the study, as well as providing an
opportunity to study gene-environment interactions).
In addition, there are other, equally important factors
associated with conducting such a study in Galicia,
such as the relatively high incidence of colorectal
cancer in its population, and the availability of
subject families (which are frequently large families
that live in the same city or town).
All of these factors have induced the
development of several recent research projects on
cancer in Galicia during the last years: “A Pilot
Study of colorectal cancer in Galicia, Spain”, funded
by the U. S. National Cancer Institute (NCI) for the
period 2004-2006, a “Colorectal Cancer Thematic
Network in Galicia”, funded by the XUNTA de
Galicia for the period 2005-2006 and a “Colorectal
Cancer Research Network in Galicia”, funded by the
XUNTA de Galicia for the period 2006-2008/9.
Within the framework of these projects, the idea of
developing an IS to improve the development of
multicenter epidemiologic studies, was jointly raised
in 2005 by the Universitary Hospital Complex Juan
Canalejo of A Coruña and the Medical Computing
and Radiological Diagnosis Center (IMEDIR
Center) of the University of A Coruña, and funded
by the XUNTA de Galicia for the period 2005-2008.
3 OBJECTIVES
The aim of the this work consists in developing a
secure IS for the achievement of multicenter
epidemiologic studies on cancer that allows 1)
collecting, storing and editing medical data from
different hospital centers, and 2) reusing the
compiled data, by means of their integration with
data from other systems in order to carry out studies
on a larger set of data.
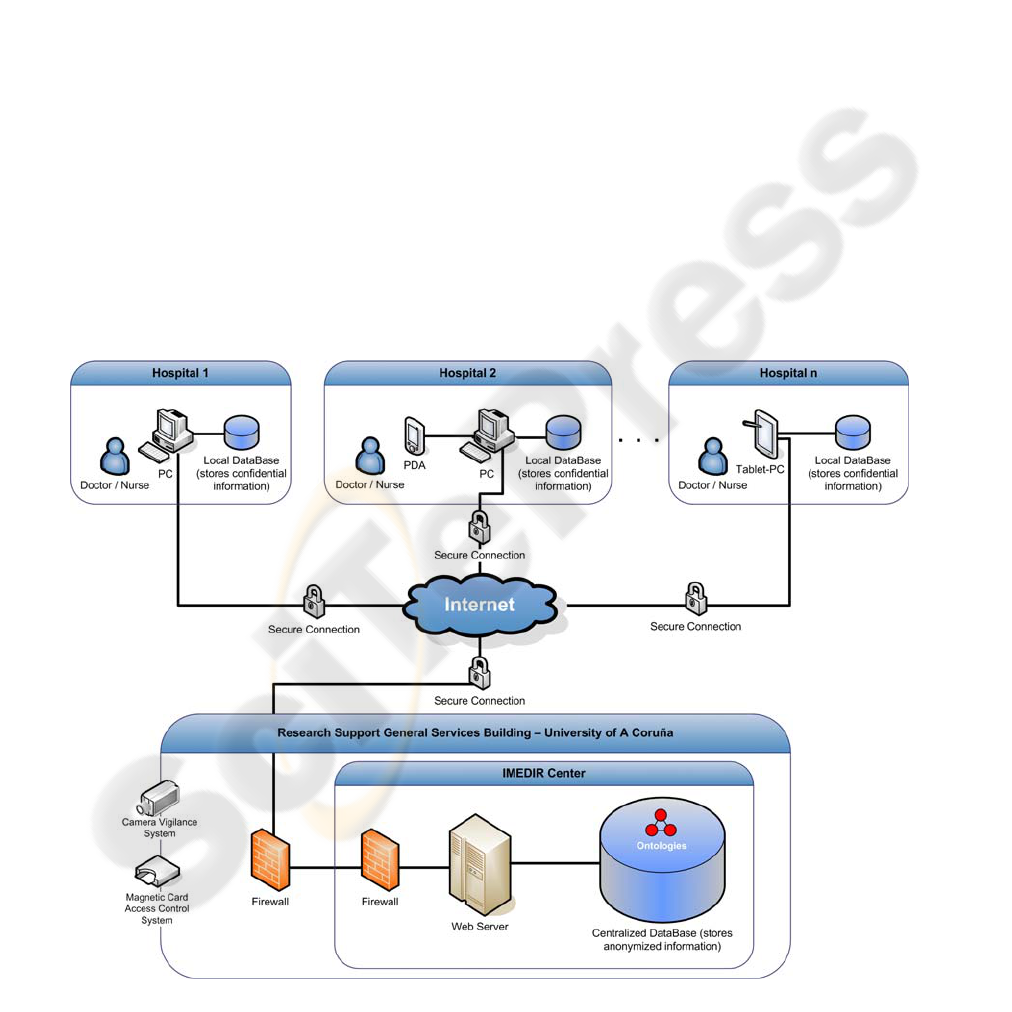
Figure 1: Physical design of the Information System.
HEALTHINF 2008 - International Conference on Health Informatics
134

4 METHODS
The development of the system was based on the
Unified Software Development Process (USDP,
Jacobson, 1999). During the Software Requirements
Specification phase (SRS), we used prototyping
techniques and interviewed many medical experts in
different hospitals. This allowed us to detect with
great detail the system requirements and to define
the interfaces according to the preferences of the
end-users.
The central system, located in the IMEDIR
Center, was developed on the J2EE platform in order
to offer more integration possibilities and follows
the architectonic patterns Model-View-Controller
(MVC) and Layers. The software for the data
compilation devices used in hospitals was developed
using the .NET Compact Framework, which
simplifies application development on smart devices
and allows to develop a very friendly user interface.
The physical design of the system is shown in the
Figure 1.
4.1 Distributed Data Collection
We studied and discussed various data collection
alternatives (Tablet PCs, PDAs, Smart Phones) and
finally opted for using Portable Digital Assistants
(PDAs), due to their characteristics of mobility, data
synchronization (e.g. with a desktop computer), data
input facilities (pen-stylus method) and an enough
size screen for a correct handling of the application
(Wiggins, 2004). The software for the PDAs was
developed on the .NET Compact Framework
because it makes it easier to build applications for
this kind of handheld devices and allows to develop
a very friendly user interface (see Figure 2). We
must consider that usability is a fundamental factor
to obtain the success of an IS, especially in medical
environments. Although at present only data
compilation software for PDAs is available, the
system is independent from the data collection
device used, so it can be easily adapted to other
devices like the previously mentioned.
The data collection software allows the
automatic validation of data, minimizing the input
errors (e.g. personal names cannot contain numerical
characters, birth dates must be past dates, etc.). In
addition, this software is able to avoid asking
questions whose answers can be inferred from
previous answers (e.g. if a patient have already
provided his birth date, he will not have to fill out
his age) and it also provides assistance to fill out the
required information: it has an on-screen help
system, shows examples of possible data inputs in
confusing questions, guides the user (e.g. the person
who makes the interview) through the questions
(because in some questionnaires the user must
follow a way or another one depending on previous
answers), avoids unasking questions by mistake or
omission, etc. All these features have been
implemented in order to decrease the length of the
medical interviews and, therefore, to reduce the cost
of the studies.
The data collected from hospitals using the
PDAs are formatted to XML and synchronized at
regular intervals by a device (PC) that is located in
each hospital and connected with the central system
at IMEDIR Center through a secure Internet
connection. A Web application, which was
developed using J2EE technology, allows the
transmission and storage of the collected data from
the PCs in hospitals to the centralized database.
Figure 2: Screenshot of the data collection software.
4.2 Centralized Data Storage
In order to maintain the data consistency, which is
fundamental in order that the studies would be valid,
and to facilitate the data exploitation, the IS stores
all the collected data in a centralized database.
Since the proposed system stores information
arriving from multiple hospitals, which could be
placed in different geographical areas (different
cities or even in different countries), and due to the
fact that the collected data belongs to the Medical
domain, in which there exists a great terminological
AN ONTOLOGY-BASED INFORMATION SYSTEM FOR MULTICENTER EPIDEMIOLOGIC STUDIES ON
CANCER
135

heterogeneity, it is crucial to use a standard
terminology for the data storage.
At present, ontologies are viewed as an ideal
solution to solve data heterogeneity problems. They
are solid vocabularies of terms and relations among
them, agreed upon by a group of people, that can
help to overcome the semantic, syntactic and
structural ambiguity that hinders communication
between different systems and data sources.
After analyzing several options, we decided to
use the NCI Thesaurus ontology in our system
(Golbeck et al., 2004), because it is published under
an open content license and it contains a broader
range of cancer-related terms than other existing
ontologies. Using this ontology as a reference, the
information coming from each hospital is annotated
by means of a common terminology in which it is
stored. This allows the use of questionnaires that can
be written in different languages or medical
terminologies depending on the geographical
location of each hospital, because the storage is done
in agreement with a common terminology.
Furthermore, this process is not restricted to the
translation of terms, but it also covers other aspects
like, for example, units of measurement or date and
hour formats.
The system is flexible enough to use other
ontology instead of the NCI Thesaurus; however, it
only supports using one ontology at a time.
4.3 Remote Data Editing
In the proposed system, data editing refers to the act
of modifying or deleting incorrect information from
patients and/or their relatives, or storing new
information that was not known before. This may be
required by the medical staff 1) before the data have
been transmitted from the hospital to the centralized
database, or 2) when the data are already stored in
the centralized database. In the following, both kinds
of data edition are described.
The first case lies in the edition of data that are
still stored in the hospitals. They are data that have
not been transmitted to the centralized database
because they are incomplete, or because they have
not been reviewed yet. The user must authenticate
himself to the system and carry out the appropriate
changes. This is a usual kind of data edition.
The second case refers to the edition of data that
have already been sent from the hospital to the
centralized database. In this case, the user works
directly against the centralized database. This option
should be used only in exceptional cases, because it
consists of modifying information that is already
assumed to be complete and reviewed. As a safety
measure that prevents the system about the loss of
important information due to user errors or about
possible attacks from outside, this kind of edition
requires, in addition to the authentication of the
doctor or nurse who wishes to edit the data, the
permission of the system administrator, who is
located in the IMEDIR Center.
4.4 Data Integration Capabilities
The data collected during the achievement of an
epidemiologic study has a great value, both because
of the difficulty and cost (in time and economic) of
this process and due to its great potential of
reusability by means of its integration with data
from other sources, which allows to carry out new
studies with a larger amount of data.
Nevertheless, the data gathered during an
epidemiologic study are rarely reused after its
finalization. This is mainly due to that traditional
storage supports (e.g. paper) are used, as well as
specific storage formats and terminologies which
cause that the reusability of the collected data by
means of the integration with data coming from
other studies is not worthwhile.
By means of the last advances in ICT, we have
provided our system with capabilities of information
reusing and integration.
The main objective of data integration is to
provide ways to unify the information from several
distributed, heterogeneous and autonomous data
sources (e.g. information systems, databases, XML
files, etc.). An integrated view must be able to
describe the various data sources and their
interrelation, overcoming the syntactic, structural
and semantic heterogeneity problems. All of this,
with the aim of automating the process of getting
data from various resources, instead of having to
manually request data from them and then combine
the results.
However, as it is explained in Chou, 2005,
Information integration is not a trivial task. Data are
usually stored in relational databases, and it is often
the case that only the creators of the databases
understand the semantic meaning of columns in each
table. Therefore, it is difficult for a user (or a
system) to integrate the data from the databases with
data from other sources without first understanding
how they are structured, or without being explicitly
told from which columns to retrieve information.
The presented system allows 1) accessing
through the Internet to the information collected
during an epidemiologic study on colorectal cancer
HEALTHINF 2008 - International Conference on Health Informatics
136
in Galicia, Spain and 2) requesting information from
any set of information sources which have been
mapped to an ontology from the cancer domain. In
the following, we will describe how these two
functionalities were achieved.
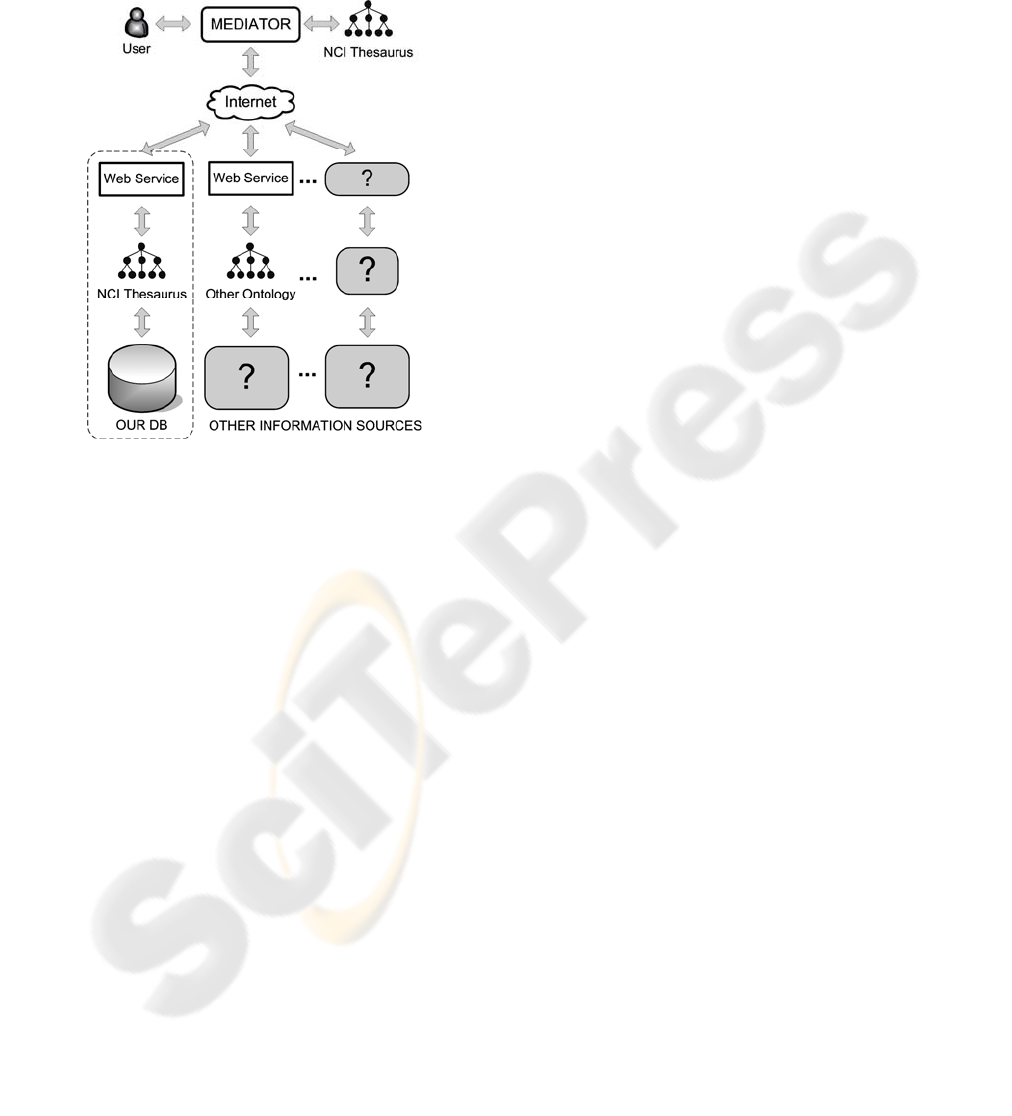
Figure 3: Data integration capabilities of the IS.
4.4.1 Making the Collected Data Publicly
Accessible
In order to make our information publicly accessible
through the Internet, we have opted for a solution
that uses ontologies and Web services.
Ontologies are useful to support the integration
of data from multiple repositories (Jakoniene &
Lambrix, 2005, Perez-Rey et al., 2005, Stevens et
al., 2000), and some of the current Integration
Information Systems incorporate ontology-related
knowledge (e.g. Deray & Verheyden, 2003, Alexiev
et al. 2005). The proposed system uses ontologies to
resolve the semantic conflicts that usually hinder
integration data by using the NCI Thesaurus
ontology as a reference vocabulary, mapping the
columns of the centralized database with terms from
that ontology.
On the other hand, Web services provide
loosely-coupled, language-neutral, and
platform-independent ways of linking applications
across the Internet. Our system is designed to allow
remote queries written in the terminology of the NCI
Thesaurus ontology, and to answer these queries
through the Internet. The elements of the IS that
provide this functionality are represented in the
Figure 3 into a dotted area.
4.4.2 Integrating Information from other
Sources
Instead of having to manually request data from
various data sources and then combine the results,
our system is also prepared to automate this process.
We have implemented this functionality on the basis
of one of the ontology-based information integration
approaches (the “Single Ontology Approach”),
proposed by Wache et al. in 2001.
In this process (see Figure 3), the IS acts as a
“Mediator” that 1) receives a request (query) from
the user, 2) processes the query and ask a set of data
sources that have been prepared to be accessed
through Web services, and 3) puts together all the
results from the data sources and returns the
combined result to the user.
At the moment, this functionality is in testing
phase, and it only works well with information
sources that are available to be queried through a
Web service by means of the terminology of the NCI
Thesaurus. However, the preliminary results have
been very satisfactory, and our intention is to
continue improving this functionality to reach the
most general and automatic behaviour possible.
4.5 Security Issues
The development of this IS also considers the
security requirements imposed by Spanish law and
Galician regulations, which are among the most
restrictive European legislations, as well as the
United States Safe Harbor Agreement. Under the
Spanish legislation in force, the medical data in this
IS are considered sensible data and specially
protected, thus the safety measures acquire special
importance. The data collection is made by the
medical staff in hospitals, that is authorized by the
law for the managing and processing of data about
personal health (LOPD 15/1999, 1999). The used
PDAs have an integrated biometric fingerprint
reader, which provides security access to the
personal data stored in the device and univocally
identifies the user, according to the measures
arranged by the law (RD 994/1999 , 1999).
In order to transfer the collected data from the
PDAs to the PC, both devices are connected by
cable, and the doctor or nurse is authenticated in the
PC by means of a cryptographic smart card of the
Galician Service of Health (SERGAS). In this smart
card there is stored a digital certificate issued by the
Fábrica Nacional de Moneda y Timbre (FNMT), a
certifier authority recognized by the Spanish state
that univocally identifies the user who uses it. All
AN ONTOLOGY-BASED INFORMATION SYSTEM FOR MULTICENTER EPIDEMIOLOGIC STUDIES ON
CANCER
137

accesses to the data are totally monitored and
registered, and it is registered for the later
accomplishment of audits (Wei et al., 2006). When
transferring the data to the PC, a dissociation process
is made in which the personal character data
necessary for the medical personnel to identify the
subject (e.g. the number of clinical history of the
patient) and the genetic-environmental data required
by the epidemiologic study, that will be transferred
later to the centralized database located in the
IMEDIR Center, are separated. In this process the
patient’s identity is dissociated of its clinical data,
which are anonimous under a numerical code, and
both are stored in the PC in a separated way. The
relation between both data types is stored in a file
which will only be accessed from the hospital.
The transmission of the anonimous data from the
PCs in the hospitals to the centralized database is
made over the Internet and through a Web
application, in which the user is authenticated by
means of the same mechanism that he/she uses when
is connected to the PC in the hospital, that is, by
means of the digital certificate that resides in the
cryptographic smart card of the SERGAS. To assure
the safety of the data during the transmission, all the
data tranfers are carried out on encrypted
connections using the Secure HyperText Transfer
Protocol (HTTPS) over the Secure Socket Layer
(SSL). To incorporate this method, a server security
certificate needs to be configured on the server, so
these technologies and protocols use public/private
key technologies (Cooper et al., 2006, Bourasa et al.,
2005). Likewise, the IMEDIR Center has an
architecture of double firewall (see Figure 1), in
which the first firewall of the building limits the
access to prevent external generic attacks, whereas
the second firewall, placed inside the IMEDIR
Center, restricts the access to the Web application by
IP address, so that only those IPs that have been
authorized (the PCs of the hospitals) can connect
with the application to transfer the data. This system
has been chosen as the most suitable due to the fact
that resting on the HTTPS protocol the development
of Web services is quite simple, and they can take
advantage of the firewall safety systems without
need to change the filter rules (Stanton, 2005).
Although the data stored in the IMEDIR Center
are anonymous, the peculiar characteristics of some
gathered families might allow their identification,
what makes necessary to maintain a high level of
security at all time. With the purpose of providing an
environment as safe as possible in the IMEDIR
Center, it has several physical safety measures, like
security cameras that provide 24-hour video
vigilance and the use of cryptographic smart cards to
control the access to the building.
5 RESULTS
The presented IS allows to carry out epidemiologic
studies on cancer that require less time for
interviews with patients and present less errors in the
compiled data, while guaranteeing the integrity of
these data. The system satisfies the special demands
of modern medical information systems, such as
security and interoperability. The use of this IS
allows to save time and money, and increase the
reliability of the performed studies. It also allows us
to integrate our data with other information systems,
through the Internet, and as such carry out new
studies with a larger amount of data (this is
particularly important in cancer studies).
This system has been successfully applied in the
execution of the “Pilot Study of Colorectal Cancer in
Galicia, Spain”, financed by the U.S. National
Cancer Institute.
6 CONCLUSIONS
The study of multifactorial diseases such as cancer
requires a large amount of data that need to be
compiled, stored and analysed, and from which new
information must be extracted. In addition, reusing
these data in other similar studies would provide
great benefits.
Information and Communication Technologies
can contribute significantly to this task thanks to the
development of Information Systems such as the
presently proposed one. This system, allows
collecting, storing and editing medical data from
different hospitals in a secure manner, and reusing
the compiled data by means of their integration with
data from other information sources with the
purpose of carrying out studies on a larger set of
data. The usefulness of this IS has been
demonstrated during the development of a real
epidemiologic study of colorectal cancer in Galicia,
Spain.
7 FUTURE DIRECTIONS
In the following, some of the ideas that could help to
improve the proposed IS are presented:
HEALTHINF 2008 - International Conference on Health Informatics
138

With regard to the data integration capabilities of
the system, we are thinking about developing an
advanced mechanism to automatically retrieving
information from sources whose information has
been prepared to be accessed. This mechanism could
be based on a set of intelligent semantic agents that
would interoperate with the various information
sources through the terminology of existing
ontologies. This would allow us automatically
retrieve and integrate a huge amount of data from
other studies that we would analyse in order to make
progresses in the treatment of pathologies like
colorectal cancer.
It also could be useful to provide the system with
data mining techniques (Tan, Steinbach & Kumar,
2006). These techniques could be deployed to scour
the large amount of epidemiologic data compiled in
order to find novel and useful patterns that might
otherwise remain unknown, and they would also be
useful to predict the outcome of future observations.
All this would help to decrease the incidence of
diseases like the cancer, and to improve its
prevention and treatment.
ACKNOWLEDGEMENTS
This work was partially supported by the Spanish
Ministry of Education and Culture (Ref TIN2006-
13274) and the European Regional Development
Funds (ERDF), grant (Ref. PIO52048) funded by the
Carlos III Health Institute, grant (Ref. PGIDIT 05
SIN 10501PR) from the General Directorate of
Research of the Xunta de Galicia and grant (File
2006/60) from the General Directorate of Scientific
and Technologic Promotion of the Galician
University System of the Xunta de Galicia. The
work of José M. Vázquez is supported by an FPU
grant (Ref. AP2005-1415) from the Spanish
Ministry of Education and Science.
REFERENCES
Alexiev, V., Breu, M., de Bruijn, J., Fensel, D., Lara, R.,
Lausen H., 2005. Information Integration with
Ontologies – Experiences from an Industrial
Showcase. John Wiley & Sons.
Bourasa, C., Gkamasa, A., Naveb, I., Primpasa, D.,
Shanib, A., Sheoryb, O., Stamosa, K., Tzruyac, Y.,
2005. Application on demand system over the Internet.
Journal of Network and Computer Applications. 28(3),
209-232.
Cooper, C. J., Cooper, S. P., Junco, D. J., Shipp, E. M.,
Whitworth, R., Cooper S. R., 2006. Web-based data
collection: detailed methods of a questionnaire and
data gathering tool. Epidemiologic Perspectives &
Innovations. Volume 3.
Chou, H. H., 2005. BioDig: Architecture for Integrating
Heterogeneous Biological Data Repositories Using
Ontologies. Thesis. Massachusetts Institute of
Technology.
Deray, T., Verheyden, P., 2003. Towards a Semantic
Integration of Medical Relational Databases by Using
Ontologies: A Case Study. OTM Workshops, LNCS
2889, 137-150. Springer-Verlag.
Golbeck, J., Fragoso, G., Hartel, F., Hendel, J., Parsia, B.,
2004. The National Cancer Institute’s Thesaurus and
Ontology. Journal of Web Semantics, Vol. 1, issue 1.
Jacobson, I., Booch, G., Rumbaugh, J., 1999. The Unified
Software Development Process. Addison Wesley.
Jakoniene, V., and Lambrix, P., 2005. Ontology-based
Integration for Bioinformatics. Proceedings of VLDB
Workshop on Ontologies-based techniques for
DataBases and Information Systems (ODBIS2005),
Trondheim, Norway.
Organic Law 15/1999 of December 13, 1999 on the
Protection of Personal Data.
Royal Decree 994/1999 of June 11, 1999.
Perez-Rey, D., Maojo, V., Garcia-Remesal, M., Alonso-
Calvo, R., Billhardt, H., Martin-Sanchez, F., Sousa,
A., 2005. ONTOFUSION: Ontology-Based
Integration of Genomic and Clinical Databases.
Computers in Biology and Medicine.
Stanton, R., 2005. Securing VPNs: comparing SSL and
IPsec. Computer Fraud & Security. 2005(9), 17-19.
Stevens, R., Baker, P., Bechhofer, S., Ng, G., Jacoby, A.,
Paton, N., Goble, C., Brass, A., 2000. TAMBIS:
Transparent access to multiple bioinformatics
information sources. Bioinformatics, 16(2), 184-186.
Tan, P., Steinbach, M. and Kumar, V., 2006. Introduction
to Data Mining. Addison Wesley.
Wache, H., Vogele, T., Visser, U., Stuckenschmidt, H.,
Schuster, G., Neumann, H., Hiibner, S., 2001.
Ontology-based integration of information - a survey
of existing approaches. Proceedings of the
International Workshop on Ontologies and
Information Sharing, 108-117.
Wei, J. C., Valentino, D. J., Bell, D. S., Baker, R. S., 2006.
A Web-based telemedicine system for diabetic
retinopathy screening using digital fundus
photography. Telemed J E Health, 12(1):50-7.
Wiggins, R.H., 2004. Personal digital assistants. Journal
of Digital Imaging. 17(1):5-17.
AN ONTOLOGY-BASED INFORMATION SYSTEM FOR MULTICENTER EPIDEMIOLOGIC STUDIES ON
CANCER
139
