
TOWARDS ON-DEMAND BIOMEDICAL KNOWLEDGE
EXTRACTION
Vincenzo Lanza
Anesthesia Department, Buccheri La Ferla Hospital, Fatebenefratelli, Palermo, Italy
M. Ignazia Cascio
Sicilian Center for Training and Research in Public Health (CEFPAS), Caltanissetta, Italy
Chun-Hsi Huang
Dept. of Computer Science and Engineering, University of Connecticut, Storrs, CT 06269, USA
Keywords:
Knowledge Grids, Biomedical Computing.
Abstract:
This paper outlines a UMLS-compatible distributed genomic semantic network. The system aims at providing
cooperative reasoning on distributed genomic information, complying with the UMLS concept representation,
from distributed repositories. The distributed semantic network has currently incorporated most of the 871,584
concepts (named by 2.1 million terms) of the 2002 version UMLS Metathesaurus, with inter-concept relation-
ships across multiple vocabularies and concept categorization supported. Modern information and compute
infrastructure is incorporated to allow seamless access to geographically dispersed users.
1 INTRODUCTION
The complete sequencing of numerous genomes has
stimulated new cross-domain and cross-discipline re-
search topics. Computationally, researchers have
been exploring the massive genomic and proteomic
information, attempting to generate new hypotheses
for gene/protein functions, as well as novel targets for
the development of insecticides, antibiotics, antivi-
ral drugs, and health related drugs. Semantically, re-
searchers study biological information from individ-
ual (clinical practice) to the population level (social
health-care), as well as the infrastructure for high-
performance, automated integration and analysis of
these information, in an attempt to better individual
and public health care.
It is crucial in most of these novel researches
that the massive genomic data produced are well rep-
resented so that useful biological information may
be efficiently extracted. A useful tool for effec-
tive knowledge representation is the semantic net-
work system (Lee et al., 2003). A semantic network
is a conceptual model for knowledge representation,
in which the knowledge entities are represented by
nodes (or vertices), while the edges (or arcs) are the
relations between entities (Cercone, 1992; Fahlman,
1982; Brachman and Schmolze, 1985; Shapiro and
The SNePS Implementation Group, 1998; Chung and
Moldovan, 1993; Surdeanu et al., 2002; Moldovan
et al., 1992; Evett et al., 1991; Stoffel et al., 1996).
A semantic network is an effective tool, serving as
the backbone knowledge representation system for
genomic, clinical and medical data. Usually these
knowledge bases are stored at locations geographi-
cally distributed. This highlights the importance of
an efficient distributed semantic network system en-
abling distributed knowledge integration and infer-
ence.
The semantic network is a key component of the Uni-
fied Medical Language System (UMLS) project initi-
ated in 1986 by the U.S. National Library of Medicine
(NLM). The goal of the UMLS is to facilitate associa-
tive retrieval and integration of biomedical informa-
tion so researchers and health professionals can use
such information from different (readable) sources
(Lindberget al., 1993). The UMLS project consists of
three core components: (1) the Metathesaurus, pro-
viding a common structure for more than 95 source
biomedical vocabularies. It is organized by concept,
which is a cluster of terms, e.g., synonyms, lexical
variants, and translations, with the same meaning. (2)
the Semantic Network, categorizing these concepts
102
Lanza V., Ignazia Cascio M. and Huang C. (2008).
TOWARDS ON-DEMAND BIOMEDICAL KNOWLEDGE EXTRACTION.
In Proceedings of the First International Conference on Health Informatics, pages 102-109
Copyright
c
SciTePress

by semantic types and relationships, and (3) the SPE-
CIALIST lexicon and associated lexical tools, con-
taining over 30,000 English words, including various
biomedical terminologies. Information for each en-
try, including base form, spelling variants, syntactic
category, inflectional variation of nouns and conjuga-
tion of verbs, is used by the lexical tools. The 2002
version of the Metathesaurus contains 871,584 con-
cepts named by 2.1 million terms. It also includes
inter-concept relationships across multiple vocabular-
ies, concept categorization, and information on con-
cept co-occurrence in MEDLINE.
2 THE PILOT SYSTEM
We are currently developing a UMLS-compatible dis-
tributed genomic semantic network. This system aims
at providing cooperative reasoning on distributed
genomic information, complying with the UMLS
concept representation, from distributed repositories.
Representative inference rules (path-based) and com-
mands (SNePS-like (Moldovan et al., 2003)) are
briefed in Appendices A and B.
The infrastructure of the cooperative software
components is extended from the TROJAN system
(Lee et al., 2004; Lee and Huang, 2004). The pi-
lot system emphasizes the task-based and message-
driven model to exploit parallelism at both task and
data levels. The system also features multi-threading
and task migration to support communication latency
hiding and load balancing, respectively. In the task
model, queries are decomposed into tasks and dis-
tributed among processors for execution. When a task
is completed, a message is generated to either spawn
new tasks or trigger further processing, depending on
the property and current status of the task. This pro-
cess is carried out by two collaborating components:
the host system and the slave system. The host system
interacts with users and processes information for the
slave system, while the slave system executes com-
pute tasks.
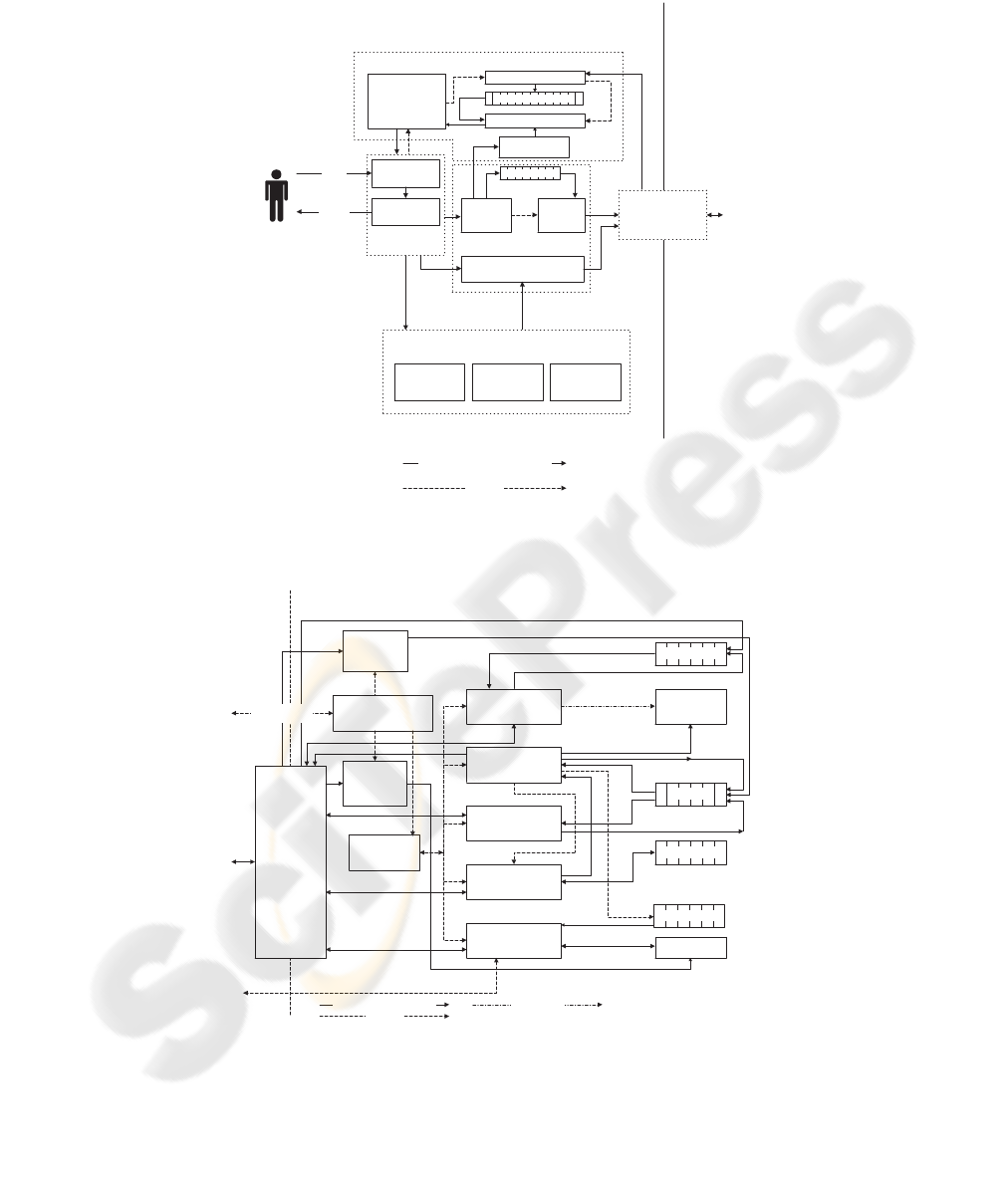
The host system is composed of the following
major components. The language front-end inter-
acts with the user and decomposes the commands
into either knowledge or tasks. All the preprocessing
and distributing are carried out in the command pro-
cessing module. The object-oriented packing module
is the communication channel between processors.
When the slave module finishes a query, the answer
messages are then sent back to the host answer pro-
cessing module of the host system to be merged into
a final inference conclusion. Some knowledge is kept
in the host knowledge base for simple queries. Fig. 5
illustrates the host system.
The major components comprising the slave sys-
tem are as follows. The shared knowledge manage-
ment module stores and exchanges knowledge in the
shared knowledge base. The task execution module
is the kernel of task execution. Several sub-modules
are embedded in the task execution module, including
the kernel message module, the task execution engine,
and the load balancing module, etc. The duplicate
checking module records the answers that have been
reached to save repeated executions. The slave sched-
uler schedules task execution and swapping. The
object-oriented packing system is similar to that of the
host. The slave system is depicted in Fig. 6.
Commands in our semantic network system are
generally categorized into three groups: (1) network
building (e.g.
build
and
assert
, etc.), (2) inferenc-
ing (e.g.
find
,
findassert
, etc.) and (3) others (e.g.
nodeset operation commands
, etc.). Commands
in groups (1) and (2) usually need to communicate
with slave PEs, while those in (3) are answered di-
rectly inside the host module. Our system provides
three commands,
build
,
assert
and
add
, to con-
struct the semantic network. The syntax of these com-
mands are listed below:
•
build
: (
build
{relation nodeset}
∗
)
•
assert
: (
assert
{relation nodeset}
∗
context-
specifier)
•
add
: (
add
{relation nodeset}
∗
context-specifier)
For example, the command
(assert member Saccharomyces-cerevisiae
class yeast)
defines the concept “Saccharomyces-cerevisiae
is yeast”. In the system, two base nodes
Saccharomyces-cerevisiae
and
yeast
are
generated by the command. The molecular node
M1
(index depending on the current knowledge base
state) is generated by the system, where “!” stands for
the “assertion” concept. Two forward links
member
and
class
are defined by the user, two reverse links
member-
and
class-
, indicated by dash lines, are
generated automatically. Hierarchical concepts can
be constructed similarly by following the links of
“subclass-” and “supclass”.
To sum up, the network building commands put a
node into the network with an arc labeled relation to
each node in the following nodeset, and returns the
newly built node. An attempt to build a currently ex-
isting node will immediately return such an existing
node.
build
creates an unasserted node unless an as-
serted node exists in the network with a superset of the
relations of the new node, in which case the new node

is also asserted.
assert
is just like
build
, but creates
the node with assertion.
add
acts like
assert
, but in
addition triggers forward inference. relation has to be
a unit-path and non converse. The converse relation
relation- that connects each node of the nodeset to the
built node is constructed implicitly by the system.
A heuristic approach is used to partition the se-
mantic network to get around the NP-hardness of op-
timal partitioning (Cormen et al., 2000; Garey et al.,
1976). Starting from PE
1
as the initial target PE,
while a network construction command is issued, the
host sends the newly built nodes to the target PE until
a certain number of nodes have accumulated and then
cyclically shifts to the next PE as the new target.
Several (path-based) inference commands are
provided by our system, including the
find
fam-
ily (
find
,
findassert
,
findbase
,
findconstant
,
findpattern
and
findvariable
). While a query
is made, corresponding tasks are generated by the
task preprocessor under the command of the parser
in the language front end, and then stored in the
host task queue temporarily while waiting to be dis-
patched by the task distributer. These query tasks
are split according to the implied parallelism of the
command. For example, when a path-based query
is made, the command is usually in the format of
(find
{path nodeset}
∗
)
, which is equivalent to
T
n
i=1
S
f(i)
j= 1
(
find
path
i
node
i, j
), where the
T
and
S
are the nodeset intersection and union, respec-
tively. These (sub)query tasks are formed and later
dispatched.
Path-based inference is the fundamental inference
mechanism of all semantic networks. By tracing the
arcs between nodes, new knowledge can be derived.
In our system, the relation between two nodes can
be either explicit (direct arc between two nodes), or
implicit (an arc across several intermediate nodes).
The implicit relation is defined by the command
define-path
.
The command
find
, designed for path-based in-
ference queries, has the following syntax:
(
find
{path nodeset}
∗
).
This command returns a set of nodes such that each
node in the set has every specified path going from
it to at least one node in the accompanying nodeset.
When the command
(find subclass (human animal))
is issued, the system answers (
M2 M3
) since
M2
and
M3
each has an edge
subclass
to either node
human
or
animal
.
The distributed semantic network has cur-
rently incorporated most of the 871,584 concepts
(named by 2.1 million terms) of the 2002 version
UMLS Metathesaurus, with inter-concept relation-
ships across multiple vocabularies and concept cate-
gorization supported.
3 BASIC INFRASTRUCTURE
Modern Grid technology represents an emerging and
expanding instrumentation, computing, information
and storage platform that allows geographically dis-
tributed resources, which are under distinct control,
to be linked together in a transparent fashion (Berman
et al., 2003; Foster and Kesselman, 1999). The power
of the Grids lays not only in the aggregate comput-
ing ability, data storage, and network bandwidth that
can readily be brought to bear on a particular prob-
lem, but also on its ease of use. After a decade’s re-
search effort, Grids are moving out of research lab-
oratories into early-adopter production systems, such
as the Computational Grid for certain computation-
intensive applications, the Data Grid for distributed
and optimized storage of large amounts of accessible
data, as well as the Knowledge Grid for intelligent use
of the Data Grid for knowledge creation and tools to
all users.
Here we refer to the Cross-Campus (or Continent)
Computational Grid as the C
3
-Grid; and the Cross-
Campus (or Continent) Data Grid as the C
2
D-Grid.
The development of the C
3
-Grid portal focuses on
the establishment of a robust set of APIs (Application
Programming Interfaces). The implementation of C
3
-
Grid is largely based on the Globus Toolkit middle-
ware (2.2.4) The web portal is served by an Apache
HTTP Server located at the University of Connecti-
cut. The C
3
-Grid database regularly aggregates com-
pute platform statistics such as job status, backfill
availability, queue schedule, as well as production
rates. The job monitoring system provides real-time
snap shots of critical computational job metrics stored
in a database and presented to the user via dynamic
web pages. Computation jobs are classified into a few
classes, each with a pre-specified priority. Statistics
for each job class are created in a real-time manner so
as to provide intelligent management of resources.
The C
2
D-Grid adds another dimension of func-
tionality to the C
3
-Grid in terms of efficient man-
agement of the often-curated biomedical knowledge-
base. Our goal is to transparently and efficiently man-
age the biomedical knowledge-base distributed across
the participating campuses, providing access via a
uniform interface (web-portal). Basic file manage-
ment functions are available via a user-friendly and
platform-independent interface. Basic file transfer,
editing and search capabilities are available via a uni-

form interface. The logical display of files for a given
(local or remote) user is also available. Data/file mi-
gration is implemented to minimize the bandwidth
consumption and to maximize the storage utilization
rate on a per user basis.
Additional technical and configuration details in
regards to the compute and data grid infrastructure
are elided, according to reviewers’ comments and the
page limit.
4 WORKFLOW CONTROL
The design of the our workflow control toolkit over
the C
3
-Grid is largely based on the Genome Analy-
sis and Database Update system (GADU) (Pearson,
1994; Shpaer et al., 1996; Mulder, 2003; Bateman
et al., 2002; Henikoff et al., 1999; Pearl et al., 2003;
Sulakhe et al., ). GADU has successfully used Grid
resources with different architectures and software
environments like the 64-bit processors in TeraGrid
and 32-bit processors in the Open Science Grid or
DOE Science Grid
1
.
The opportunistic availability and the different
architectures and environments of these resources
make it extremely difficultto use them simultaneously
through a single common system. GADU addresses
these issues by providing a resource-independent sys-
tem that can execute the bioinformatics applications
as workflows simultaneously on these heterogeneous
Grid resources. and is easily scalable to add new Grid
resources or individual clusters into its pool of re-
sources, thus providing more high-throughput com-
putational power to its scientific applications. The
workflow control toolkit has wide applications in ge-
nomics as the interpretation of every newly sequenced
genome involves the analysis of sequence data by a
variety of computationally intensive bioinformatics
tools, the execution of result and annotation parsers,
and other intermediate data-transforming scripts.
Our toolkit will act as a gateway to the C
3
-Grid
and the C
2
D-Grid, handling all the high-throughput
computations necessary for knowledge inference and
extraction from our semantic network. Analogous to
GADU, our workflow control toolkit will be imple-
mented in two modules, an analysis server and an
update server. The analysis server automatically cre-
ates workflows in the abstract Virtual Data Language,
based on predefined templates that it executes on dis-
tributed Grid resources. The update server updates
the integrated knowledge-base with recently changed
data from participating sites
1
http://www.doesciencegrid.org
The toolkit will execute its parallel jobs simul-
taneously on different Grid resources. It expresses
the workflows in the form of a directed acyclic graph
(DAG) and executes it on a specified Grid site using
Condor-G (Frey et al., 2002). The toolkit will use the
GriPhyN Virtual Data System (Foster et al., 2002) to
express, execute, and track the results of the work-
flows that help in using the grid resources.
To sum up, this workflow controller will provide
a resource-independent configuration to execute the
workflows over the C
3
-Grid. It can submit jobs re-
motely to a resource, as long as the resource pro-
vides a Globus GRAM interface (e.g., the Jazz clus-
ter). All the transformations of a workflow are ex-
pressed as Condor submit files and a DAG using Pega-
sus. The Condor-G submits the workflow to a remote
resource using the GRAM interface and also moni-
tors the workflow. The toolkit will also automatically
manage the dynamic changes in the state of the Grid
resources using monitoring and information services
along with the authentication and access models used
at different Grids.
5 CONCLUDING REMARKS
Biomedical research increasingly relies on globally
distributed information and knowledge repositories.
The quality and performance of future computing and
storage infrastructure in support of such research de-
pends heavily on the ability to exploit these reposito-
ries, to integrate these resources with local informa-
tion processing environments in a flexible and intu-
itive way, and to support information extraction and
analysis in a timely and on-demand manner.
This paper outlines a UMLS-compatible dis-
tributed genomic semantic network. The system pro-
vides cooperative reasoning on distributed genomic
information, complying with the UMLS concept rep-
resentation, from distributed repositories. The dis-
tributed semantic network has currently incorporated
most of the 871,584 concepts (named by 2.1 mil-
lion terms) of the 2002 version UMLS Metathesaurus,
with inter-concept relationships across multiple vo-
cabularies and concept categorization supported.
The knowledge database and semantic network
are to be installed within a cross-campus data grid
framework. The knowledge inference will be decom-
posed into sub-tasks and distributed across the partic-
ipating compute nodes for computation.

ACKNOWLEDGEMENTS
The workflow control for the C
3
-Grid job execution
was based on an earlier collaborative project and dis-
cussions on GADU with Dr. N. Maltsev at the Ar-
gonne National Lab, US.
REFERENCES
Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller,
L., Eddy, S., Griffiths Jones, S., Howe, K., Marshall,
M., and Sonnhammer, E. (2002). The Pfam protein
families database. Nucleic Acids Res., 30:276–280.
Berman, F., Fox, G., and Hey, T. (2003). Grid Comput-
ing: Making the Global Infrastructure a Reality. John
Wiley & Sons.
Brachman, R. J. and Schmolze, J. G. (1985). An Overview
of the KL-ONE Knowledge Representation System.
Cognitive Sci., 9:171–216.
Cercone, N. (1992). The ECO Family. Computers Math.
Applic., 23(2-5):95–131.
Chung, S. and Moldovan, D. I. (1993). Modeling Seman-
tic Networks on the Connection Machine. Journal of
Parallel and Distributed Computing, 17:152–163.
Cormen, T. H., Leiserson, C. E., and Rivest, R. L. (2000).
Introduction to Algorithms. McGraw-Hill.
Evett, M. P., Hendler, J. A., and Spector, L. (1991). Paral-
lel Knowledge Representation on the Connection Ma-
chine. Journal of Parallel and Distributed Computing,
22:168–184.
Fahlman, S. E. (1982). NETL: A System for Representing
and Using Real-World Knowledge. The MIT Press.
Foster, I. and Kesselman, C. (1999). The Grid: Blueprint for
a New Computing Infrastructure. Morgan Kaufmann,
San Francisco.
Foster, I., Voeckler, J., Wilde, M., and Zhou, Y. (2002).
Chimera: A virtual data system for representing,
querying, and automating data derivation. In Proceed-
ings of the 14-th Conference on Scientific and Statisti-
cal Database.
Frey, J., Tannenbaum, T., Foster, I., Livny, M., and Tuecke,
S. (2002). Condor-G: a computation management
agent for multi-institutional Grids. Cluster Comput-
ing, 5:237–246.
Garey, M. R., Johnson, D. J., and Stockmeyer, L. (1976).
Some Simplified NP-Complete Graph Problems. The-
oret. Comput. Sci., 1:237–267.
Henikoff, S., Henikoff, J., and Pietrokovski, S. (1999).
Blocks+: a non-redundant database of protein align-
ment blocks derived from multiple compilations.
Bioinformatics, 15:471–479.
Lee, C.-W. and Huang, C.-H. (2004). Toward Cooperative
Genomic Knowledge Inference. Parallel Computing
Journal, 30(9-10):1127–1135.
Lee, C.-W., Huang, C.-H., and Rajasekaran, S. (2003).
TROJAN: A Scalable Parallel Semantic Network Sys-
tem. In Proceedings of the 15th IEEE International
Conference on Tools eith Artificial Intelligence, pages
219–223.
Lee, C.-W., Huang, C.-H., Yang, L., and Rajasekaran, S.
(2004). Path-Based Distributed Knowledge Inference
in Semantic Networks. Journal of Supercomputing,
29(2):211–227.
Lindberg, D., Humphreys, B., and McCray, A. (1993).
The Unified Medical Language System. Methods Inf.
Med., 32(4):281–291.
Moldovan, D., Lee, W., Lin, C., and Chung, M. (1992).
SNAP, Parallel Processing Applied to AI. IEEE Com-
puter, pages 39–49.
Moldovan, D. I., Pasca, M., Harabagiu, S., and Surdeanu,
M. (2003). Performance Issues and Error Analysis in
an Open-Domain Question Answering System. ACM
Tran. on Information Systems, 21(2):133–154.
Mulder, N.J., e. a. (2003). The InterPro Database, 2003
brings increased coverage and new features. Nucleic
Acids Res., 31:315–318.
Pearl, F., Bennett, C., Bray, J., Harrison, A., Martin, N.,
Shepherd, A., Sillitoe, I., Thornton, J., and Orengo,
C. (2003). The CATH database: an extended pro-
tein family resource for structural and functional ge-
nomics. Nucleic Acids Res., 31:452–455.
Pearson, W. (1994). Using the FASTA program to search
protein and DNA sequence databases. Methods Mol.
Biol., 24:307–331.
Shapiro, S. C. and The SNePS Implementation Group
(1998). SNePS-2.4 User’s Manual. Department of
Computer Science at SUNY Buffalo.
Shpaer, E., M., R., Yee, D., Candlin, J., Mines, R., and
Hunkapiller, T. (1996). Sensitivity and selectivity in
protein similarity searches: a comparison of Smith-
Waterman in hardware to BLAST and FASTA. Ge-
nomics, 38:179–191.
Stoffel, K., Hendler, J., Saltz, J., and Anderson, B. (1996).
Parka on MIMD-Supercomputers. Technical Report
CS-TR-3672, Computer Science Dept., UM Institute
for Advanced Computer Studies, University of Mary-
land, College Park.
Sulakhe, D., Rodriguez, A., D’Souza, M., Wilde, M., Nefe-
dova, V., Foster, I., and Maltsev, N. Gnare: automated
system for high-throughput genome analysis with grid
computational backend. J. Clini. Monit. and Comput.,
19(4).
Surdeanu, M., Moldovan, D. I., and Harabagiu, S. M.
(2002). Performance Analysis of a Distributed Ques-
tion/Answering System. IEEE Trans. on Parallel and
Distributed Systems, 13(6):579–596.

APPENDIX
Path Definition Primitives
• unitpath ::= relation.
• unitpath ::= relation-: A unit path can be either a
relation or a converse of a relation.
• path ::= unitpath: A path can be either a unitpath
or the composition of various pathes defined by
the following definition.
• path ::= (
COMPOSE
{path}
∗
)
If x
1
, ...x
n
are nodes and P
i
is a path from x
1
to
x
i+1
, then (
COMPOSE
P
1
...P
n−1
) is a path from x
1
to x
n
.
• path ::= (
KSTAR
path)
If path P is composed with itself zero or more
times from node x to node y, then (
KSTAR
P) is
a path from x to y.
• path ::= (
KPLUS
path)
If path P is a composed with itself one or more
times from node x to node y, then (
KPLUS
P) is a
path from x to y.
• path ::= (
OR
{path}
∗
)
If P
1
is path from node x to node y or P
2
is a path
from x to y or ...or P
n
is a path from x to y then
(
OR
P
1
, P
2
...P
n
) is a path from x to y.
• path ::= (
AND
{path}
∗
)
If P
1
is path from node x to node y and P
2
is a path
from x to y and ...and P
n
is a path from x to y then
(
AND
P
1
, P
2
...P
n
) is a path from x to y.
• path ::= (
NOT
path)
If there is no path P from node x to node y, then
(
NOT
P) is a path from x to y.
• path ::= (
RELATIVE-COMPLEMENT
path path)
If P is a path from node x to node y and there is no
path Q from x to y, then
(
RELATIVE-COMPLEMENT
P Q) is a path from x to
y. The situation can be seen in Fig. 1.
• path ::= (
IRREFLEXIVE-RESTRICT
path)
If P is a path from node x to node y, and x 6= y,
then (
IRREFLEXIVE-RESTRICT
P) is a path from
x to y. The situation can be seen in Fig. 2.
• path ::= (
DOMAIN-RESTRICT
(path node) path)
If P is a path from node x to node y and Q is a path
from x to node z, then
(
DOMAIN-RESTRICT
(Q z) P) is a path from x to y.
The situation can be seen in Fig. 3.
• path ::= (
RANGE-RESTRICT
path (node path))
If P is a path from node x to node y and Q is a path
from y to node z, then
(
RANGE-RESTRICT
P (Q z)) is a path from x to y.
The situation can be seen in Fig. 4.
• path ::= (path
∗
)
The definition is the same as (
COMPOSE
{path}
∗
)
X Y
P
No such Path Q
Path
Figure 1: (
RELATIVE-COMPLEMENT
P Q).
X Y
P
X Y
Path
Figure 2: (
IRREFLEXIVE-RESTRICT
P).
X
Y
P
Q
Z
Path
Figure 3: (
DOMAIN-RESTRICT
(Q z) P).
X Y Z
P
Q
Path
Figure 4: (
RANGE-RESTRICT
P (Q z)).
Grammars
hsneps commandi ::⇒ ( hpath defn commandi )
| ( hfile commandi )
| ( hdelete commandi )
| ( hmisc commandi )
| hmulti node commandi
| hsnepsul vari
hpath defn commandi ::⇒ DEFINE hunitpathi
+
| UNDEFINE hunitpathi
+
hfile commandi ::⇒ INNET “hstringi”
| OUTNET “hstringi”
hdelete commandi ::⇒ RESETNET ’T
| RESETNET ’NIL
| RESETNET
hmisc commandi ::⇒ LISP
| hdup check commandi
| hload bal commandi

|
hknowledge base commandi
hdup check commandi ::⇒ DUPCHECK ’T
| DUPCHECK ’NIL
| DUPCHECK
hload bal commandi ::⇒ LOADBAL ’T
| LOADBAL ’NIL
| LOADBAL
hknowledge base commandi ::⇒ PRIVATE hintegeri
| PRIVATE ALL
| CACHE hintegeri
| CACHE ALL
hmulti node commandi ::⇒ ( hmultiple nodesi )
| hnode commandi
hmultiple nodesi ::⇒ hnode commandi
+
hnode commandi ::⇒ ( hinference commandi )
| ( hdisplay commandi )
| ( hnet build commandi )
| ( hnodeset op commandi )
| hsnepsul vari
| hmulti dollar nodei
| hmulti hash nodei
| hnode namei
hinference commandi ::⇒ FIND
hmulti path nodeseti
| FINDASSERT
hmulti path nodeseti
| FINDCONSTANT
hmulti path nodeseti
| FINDBASE
hmulti path nodeseti
| FINDVARIABLE
hmulti path nodeseti
| FINDPATTERN
hmulti path nodeseti
hdisplay commandi ::⇒ DUMP
hmulti node commandi
| DESCRIBE
hmulti node commandi
hnet build commandi ::⇒ ASSERT
hmulti relation nodeseti
| BUILD
hmulti relation nodeseti
hnodeset op commandi ::⇒ &
hmulti node commandi hmulti node commandi
| + hmulti node commandi
hmulti node commandi
| - hmulti node commandi
hmulti node commandi
| = hmulti node commandi
hsymboli
| hmulti node commandi
hunitpathseti
| > hunitpathseti hsymboli
hsnepsul vari ::⇒ * hsymboli
hmulti dollar nodei ::⇒ hdollar nodei
| ( hdollar nodeseti )
hdollar nodeseti ::⇒ hdollar nodei
+
hdollar nodei ::⇒ $ hsymboli
hunitpathseti ::⇒ hunitpathi
| ( hmulti unitpathi )
hmulti unitpathi ::⇒ hunitpathi
+
hnode namei ::⇒ hsymboli
| hintegeri
hmulti hash nodei ::⇒ hhash nodei
| ( hhash nodeseti )
hhash nodeseti ::⇒ hhash nodei
+
hhash nodei ::⇒ # hsymboli
hquestion nodei ::⇒ ? hsymboli
hmulti relation nodeseti ::⇒ hrelation nodeseti
+
hrelation nodeseti ::⇒ hrelationi
hmulti node commandi
hmulti path nodeseti ::⇒ hpath nodeseti
+
| hpath question nodei
| hmulti path nodeseti
hpath question nodei
hpath question nodei ::⇒ hpathi hquestion nodei
hpath nodeseti ::⇒ hpathi hmulti node commandi
hpathi ::⇒ hunitpathi
| (COMPOSE hmulti pathi)
| ( KSTAR hpathi )
| ( KPLUS hpathi )
| ( OR hmulti pathi )
| ( AND hmulti pathi )
| ( NOT hpathi )
|
(RELATIVE COMPLEMENT hpathi hpathi)
|
(IRREFLEXIVE RESTRICT hpathi)
| (DOMAIN RESTRICT (
hpathi hnode namei ) hpathi)
| (RANGE RESTRICT
hpathi ( hpathi hnode namei ))
| (hmulti pathi)
hmulti pathi ::⇒ hpathi
+
hrelationi ::⇒ hunitpathi
| hunitpathi-
hunitpathi ::⇒ hsymboli
hdigiti ::⇒ [0-9 ]
hnon neg chari ::⇒ [0-9a-zA-Z ]
hintegeri ::⇒ hdigiti
+
hsymboli ::⇒ hnon neg chari
+
hstringi ::⇒ hchari
+
Software Architectures
Object-Oriented
Packing System
Host Scheduler
Task
Distributor
Knowledge Distributor
Task
Preprocessor
Post Task
Processes Pool
Language
Front-End
Lex Module
Yacc Module
Kernel Message Processor
Answer
Query
Host Module
Slave
Module
Task Queue
Command
Processing Module
Kernel Message Queue
Host Knowledge Base
Symbol Table Host Net Node Map
Host Answer Processing Module
Control Path
Data Generation/Consumption/Access Path
Kernel Message Receiver
Figure 5: Host System Software Architecture.
Slave Scheduler
Object-
Oriented
Packing
System
Duplication Checking
Module
Shared Knowledge
Module
Slave PE Main Module
Initial Knowledge
Receiver
(from Host)
Host/Remote
PEs
Shared Knowledge
Base
Initial Task
Receiver
(from Host)
Control Path
Data Generation/Consumption Path Data Access Path
Control Messages
with Host
Duplication Table
Memory Task
Queue
Load Balancing
Module
Task Execution
Module
Slave PE
Kernel Message
Module
Task Queue
Post Task
Processes Pool
Kernel Message
Queue
Figure 6: Slave System Software Architecture.
