
IMPLEMENTATION OF EPS_T2DM
Implementation of Early Prediction System for Type 2 Diabetes Mellitus
Dan Bi Kim, Ju-young Lee, Eun-jung Jang, Ji-young Lee and Taek Kim
Division of Bio-Medical Informatics, Center for Genome Science
Korea Centers for Disease Control and Prevention, Korea National Institute of Health
194, TongIl-ro, EunPyong-Gu, Seoul, Korea
Keywords: Type 2 Diabetes Mellitus, Early Prediction System, KoGES.
Abstract: This paper describes the implementation of an early prediction system for Type 2 diabetes mellitus. Type 2
diabetes mellitus is a multifactorial disease. It is not only associated with an unhealthy lifestyle but also has
a strong genetic component. Accordingly, in order to decrease an incidence rate of T2DM, it is important to
predict T2DM risk with using multifactors which are supposed to affect T2DM. We have implemented a
prediction system for T2DM, and it employs several statistical prediction models. These models are
produced by statistical analysis about cohort data of Korean Genome and Epidemiology Study (KoGES),
and include risk factors which are adequate for preventing T2DM in Korean populations. The prediction
system is written in JSF and Java, and developed into web application which is designed through object
oriented modeling. Web application of this system offers user interfaces in order to input data which is
needed for predicting risk group, select predefined prediction models, and so on. The system provides the
results which are predicted by selected models using inputted information.
1 INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a multifactorial
disease in which environmental triggers interact with
genetic variants in the predisposition to the disease.
In the last thirty years, due to rapid industrialization
and the lifestyle change, the number of Korean
patients with T2DM is on an increasing trend. In
addition, more serious problem is that some
significant number of T2DM patients do not aware
of their disease before onset it. Therefore, it is not
only important to treat T2DM, but also necessary to
predict its risk and prevent it in advance.
To prevent and control a certain disease for
specified population effectively, it is necessary to
understand epidemiological features of patients. So,
we have analyzed the data collected by cohort study
from Korea National Institute of Health (KNIH).
The primary prevention method of a disease is
prediction of its risk group or patients unaware of
their disease, and decreasing the incidence of the
disease or complications by reduction of risk factors
for that group. For that reason, we have
implemented the early prediction system for T2DM
which is named EPS_T2DM.
2 MATERIALS AND METHODS
At first, we have analyzed KoGES data to find risk
factors for T2DM, and built prediction models by
statistical analysis. Then, for constructing database
schema, input values including risk factors are
defined. Finally, the entire system is implemented as
input modules, a select model module, model
processing modules and displaying module for
results.
2.1 Building Statistical Prediction
Models
This study has been supported by community based
cohort study Ansung and Ansan, Korean Genome
and Epidemiology Study (KoGES) from KNIH. The
data for this study consists of epidemiological
information and genetic information (SNP values)
for 212 T2DM public and 472 general (non-T2DM)
public (Table 1).
254
Bi Kim D., Lee J., Jang E., Lee J. and Kim T. (2008).
IMPLEMENTATION OF EPS T2DM - Implementation of Early Prediction System for Type 2 Diabetes Mellitus.
In Proceedings of the First International Conference on Health Informatics, pages 254-257
Copyright
c
SciTePress
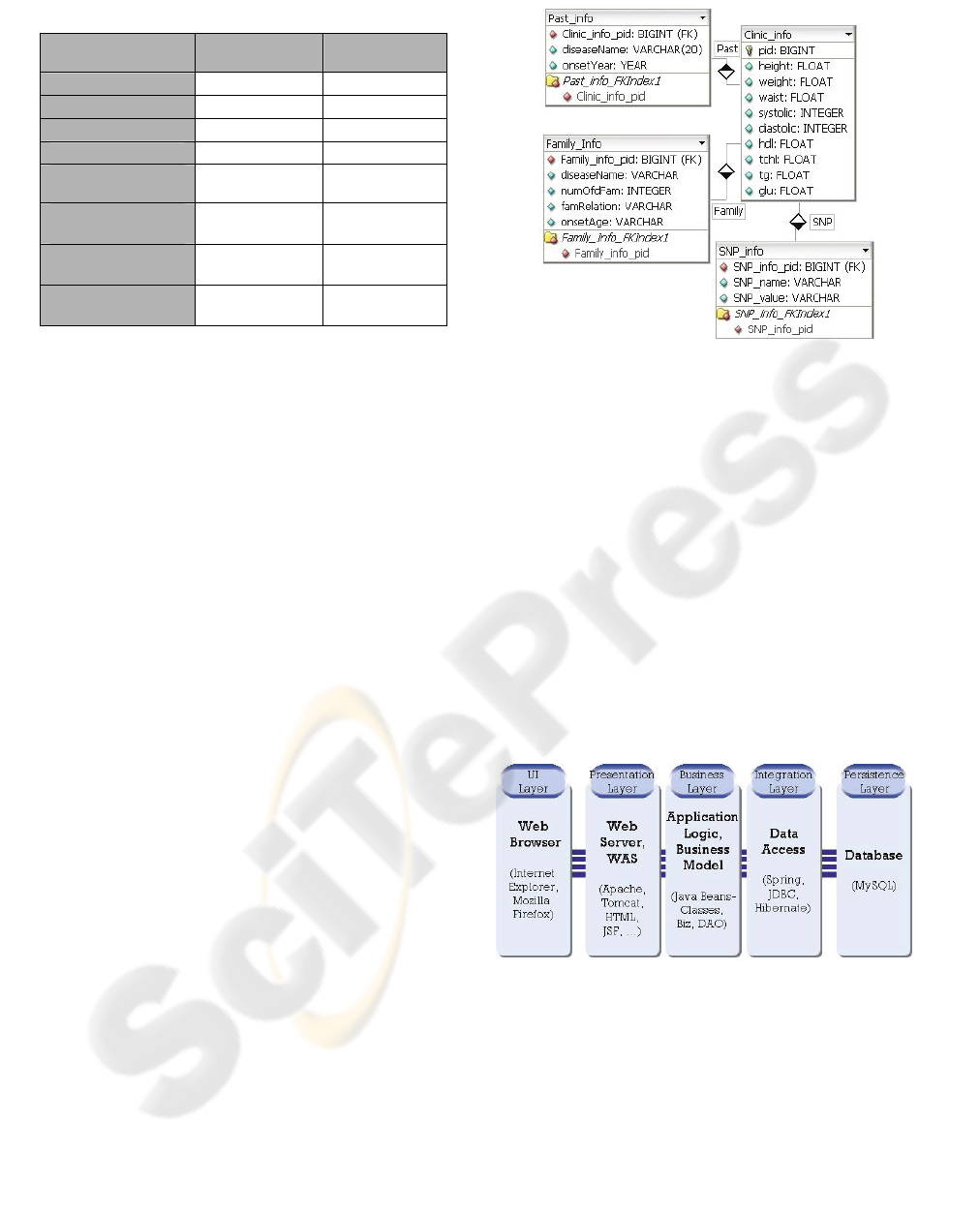
Table 1: Basic characteristics. (Means±SD).
Characteristic Diabetes Non Diabetes
Numbers
212 472
Sex(F/M)
133/79 264 / 208
Age(Years)
64.50±2.82 64.03±2.86
BMI(kg/m
2
)
25.07±3.44 23.31±3.11
Triglyceride
(mg/dl)
196.60±131.69 149.52±70.97
Fasting
glucose(mg/dl)
111.20±28.96 74.54±3.55
Total
cholesterol(mg/dl)
202.78±44.79 180.66±31.64
HDL
cholesterol(mg/dl)
44.56±10.59 44.34±9.91
The Ansung and Ansan cohort data have been
processed using the Statistical Analysis System for
Windows (Ver. 9.1, SAS institute Inc., Cary, NC,
U.S.A), and mined by several algorithms such as
QUEST, C4.5, logistic regression, SVM, and KNN
algorithm. Among data mining results, the results of
QUEST, a binary-split decision tree algorithm for
classification and data mining, is applied to
EPS_T2DM. From this result, risk factors for T2DM
are defined and prediction models are produced.
2.2 Design of Database for Input
Values
According to data mining results, risk factors for
T2DM are as follows:
Clinical information – Height and weight for
BMI, Waist circumference for abdominal
fatness, Blood pressure, Total cholesterol,
High-density cholesterol, Triglyceride, Fasting
glucose, and etc;
History of diseases information – Diabetes
Mellitus, Cerebrovascular disease and other
vascular diseases, Hypertension;
Family history of diseases information –
Diabetes Mellitus;
Genetic Information – Selected single
nucleotide polymorphisms (SNPs) in 15 genes
in Insulin pathway, 8 genes in fatty acid
binding/translocation, and 13 genes in GLUT4
translocation and 51 more genes related to
T2DM.
Each category is represented in a table, and each
factor is corresponds to each field (Figure 1).
Figure 1: Database schema for input values.
2.3 Implementing EPS_T2DM
EPS_T2DM is based on Object Oriented Modeling.
It is developed on Fedora core 6, written in HTML,
JSF, JavaScript, Java, and etc. MySQL is used as a
database to store and access data. The entire system
is implemented with Spring Framework which is a
layered Java/J2EE application framework, and then
Model-View-Controller (MVC) design pattern is
applied. MVC is an architectural pattern which
encapsulates some data together with its processing
(the model) and isolates it from the manipulation
(the controller) and presentation (the view) part.
Figure 2 illustrates the system architecture which is
composed of five layers with UI layer, 3 layers
above, and persistence layer.
Figure 2: System architecture of EPS_T2DM.
The function of EPS_T2DM consists of 4 parts:
(1) user interfaces to get input data and user’s
selection, (2) management modules of inputted data,
(3) processing and management modules of
statistical prediction models, and (4) interfaces to
offer prediction results. JSF is used to implement
user interfaces, and XML is employed to manage
input data, SNP information, and prediction models.
IMPLEMENTATION OF EPS_T2DM
255
3 RESULTS
EPS_T2DM is implemented as a web application. It
offers interfaces in order to input SNP values and
epidemiology data which include clinical
information, history of diseases information and
family history of diseases information. After getting
input data, the system shows prediction models to
users, and users can select them. The system applies
selected models to user’s input values, and displays
prediction results that are represented whether or not
a person corresponding to the inputted data belongs
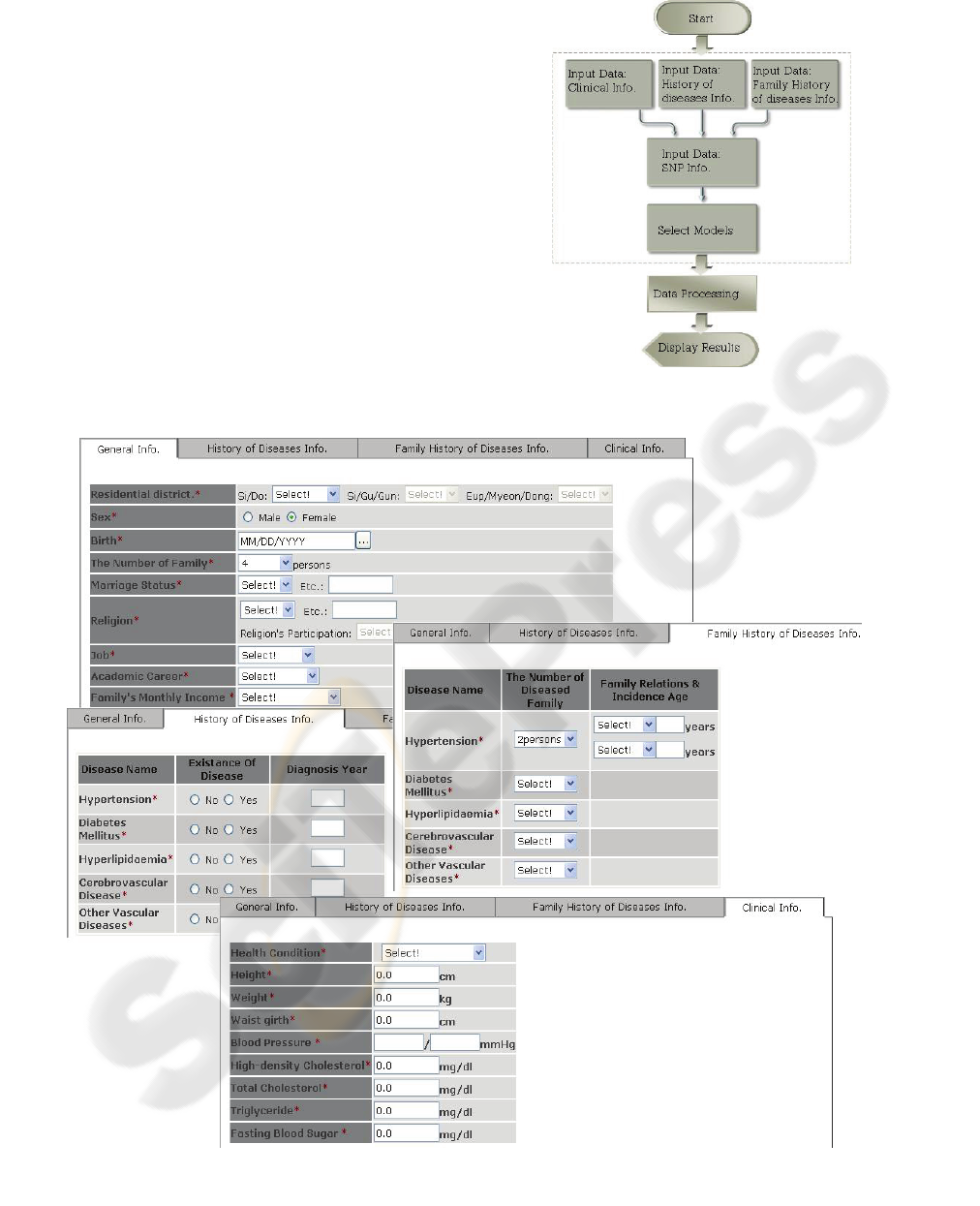
to risk group of T2DM. Figure 3 gives a flow chart
of this system.
The web interfaces consist of epidemiology
information input page with tabbed pane (Figure 4),
SNP values input page, selecting models page,
displaying results page, and so on.
Figure 3: Web application’s flow chart of EPS_T2DM.
Figure 4: The snapshots of EPS_T2DM’s web application. The interface for inputting epidemiology data which is defined
as risk factors of T2DM and other additive information such as sex, birth, etc.
HEALTHINF 2008 - International Conference on Health Informatics
256

4 DISCUSSION
In many cohort studies, various T2DM predicting
models have been developed to guide intervention
and inform health policy. Most of these studies
tested models that only used personal information
and clinical variables, not including genetic
information. However, since development of T2DM
is influenced by a complex interaction between
genetic and environmental factors, genetic
information is also needed to develop the prediction
models for T2DM.
In this study, we implemented the system to
predict the T2DM risk group with applying T2DM
risk factors and statistical prediction models that
include clinical and genetic information. The risk
factors and prediction models are base on Ansung
and Ansan, KoGES data and decision tree learning
method, and these can be updated or changed by
based data or analysis methods. EPS_T2DM is
developed as object-oriented program, so it is easy to
extended and enhance the system.
Our next step will be to expand and improve the
system. This includes followings:
The betterment of statistical models. This
means improving accuracy or building new
prediction models by analyzing other data or
applying other data mining methods
The enhancement of web application for
uploading or processing massive data sets.
REFERENCES
Zimme P, Alberti KG, Shaw J. (2001). Global and societal
implications of the diabetes epidemic. In Nature, 414,
782-787.
Ju-young Lee, Eun-jung Jang, Ji-young Lee, Kyung-soo
Oh, Hyun-woo Han (2007). PTPN1 Gene: The
haplotype-based interaction with Type 2 Diabetes. In
ISMB 2007, Poster 182.
Peter W.F. Wilson, James B. Meigs, Lisa Sullivan,
Caroline S. Fox, David M. Nathan, Ralph B.
D’Agostino. (2007). Prediction of Incident Diabetes
Mellitus in Middle-aged Adults. In Archives of
Internal Medicine, 167, 1068-1074.
The Diabetes Prevention Program Research Group.
(2005). Strategies to Identify Adults at High Risk for
Type 2 Diabetes. In Diabetes Care, 28, 138-144.
Alok Bhargava. (2003). A longitudinal analysis of the risk
factors for diabetes and coronary heart disease in the
Framingham Offspring Study. In Population Health
Metrics, 1:3.
Enzo Bonora, Stefan Kiechl, Johann Willeit, Friedrich
Oberhollenzer, Georg Egger, James B. Meigs,
Riccardo C. Bonadonna, Michele Muggeo. (2004).
Population-Based Incidence Rates and Risk Factors for
Type 2 Diabetes in White Individuals. In Diabetes, 53,
1782-1789.
Kyong Soo Park. (2004). Prevention of Type 2 diabetes
mellitus from the viewpoint of genetics. In Diabetes
Research and Clinical Practice, 66S, S33-S35.
IMPLEMENTATION OF EPS_T2DM
257
