
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT
OF EFFICIENT KNOWLEDGE DISCOVERY FROM
MULTILEVEL CLINICAL AND GENOMIC DATASETS
Manolis Tsiknakis, Stelios Sfakianakis
Institute of Computer Science, Foundation for Research and Technology - Hellas
GR-71110 Heraklion, Crete, Greece
Stefan Rueping
Fraunhofer IAIS, Schloss Birlinghoven, 53754 St. Augustin, Germany
Oswaldo Trelles
Computer Architecture Dept., University of Malaga, Bulevar Louis Pasteur 45, 29010, Malaga, Spain
Thierry Siestang
Swiss Institute of Bioinformatics, Bâtiment GénopodeCH-1015 Lausanne, Switzerland
Brecht Claerhout
Custodix NV, Verlorenbroodstraat 120b14, Merelbeke, Belgium
Vasiilis Virvilis
Biovista S.A. 34 Rodopoleos Street, Ellinikon, Athens 1677, Greece
Keywords: Ontology-based Biomedical Database Integration, Semantic Mediation, Ontologies, Post-genomic Clinical
Trials, Service Oriented Architectures.
Abstract: This paper presents the architectural considerations of the Advancing Clinico-Genomic Trials on Cancer
(ACGT) project aiming at delivering a European Biomedical Grid in support of efficient knowledge
discovery in the context of post-genomic clinical trials on cancer. Our main research challenge in ACGT is
the requirement to develop an infrastructure able to produce, use, and deploy knowledge as a basic element
of advanced applications, which will mainly constitute a Biomedical Knowledge Grid.
Our approach to offer semantic modelling of available services and data sources to support high level
services and dynamic services for discovery and composition will be presented. In particular, ontologies
and metadata are the basic elements through which Grid intelligence services can be developed, and the
current achievements of the project in this domain will be discussed.
1 INTRODUCTION
Life sciences are currently at the centre of an
informational revolution. Dramatic changes are
being registered as a consequence of the
development of techniques and tools that allow the
collection of biological information at an
unprecedented level of detail and in extremely large
quantities.
The nature and amount of the information now
available open directions of research that were once
in the realm of science fiction. Pharmacogenomics
(Roses, 2000), diagnostics (Sotiriou, 2007) and drug
279
Tsiknakis M., Sfakianakis S., Rueping S., Trelles O., Siestang T., Claerhout B. and Virvilis V. (2008).
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT OF EFFICIENT KNOWLEDGE DISCOVERY FROM MULTILEVEL CLINICAL AND
GENOMIC DATASETS.
In Proceedings of the First International Conference on Health Informatics, pages 279-287
Copyright
c
SciTePress

target identification (Schuppe-Koistinen, 2007) are
just a few of the many areas that have the potential
to use this information to change dramatically the
scientific landscape in the life sciences.
During this informational revolution, the data
gathering capabilities have greatly surpassed the
data analysis techniques. If we were to imagine the
Holy Grail of life sciences, we might envision a
technology that would allow us to fully understand
the data at the speed at which it is collected. Ideally,
we would like knowledge manipulation to become
tomorrow the way goods manufacturing is today:
highly automated, producing more goods, of higher
quality and in more cost effective manner than
manual production. It is our belief that, in a sense,
knowledge manipulation is now reaching its pre-
industrial age. The explosive growth in the number
of new and powerful technologies within proteomics
and functional genomics can now produce massive
amounts of data but using it to manufacture highly
processed pieces of knowledge still requires the
involvement of skilled human experts to forge
through small pieces of raw data one at a time. The
ultimate challenge in coming years, we believe, will
be to automate this knowledge discovery process.
This paper presents a short background section
discussing the urgent needs faced by the biomedical
informatics research community, and very briefly
describes the clinical trials upon which the ACGT
project is based for both gathering and eliciting
requirements and also for validating the
technological infrastructure designed. It continues
with a presentation of the initial ACGT architecture
defined, and presents its layers and key enabling
services.
2 POST-GENOMIC CLINICAL
TRIALS
In ACGT we focus in the domain of clinical trials on
cancer. Cancer, being a complex multifactorial
disease group that affects a significant proportion of
the population worldwide, is a prime target for
focused multidisciplinary efforts using currently
available novel, high throughput and powerful
technologies. Exciting new research on the
molecular mechanisms that control cell growth and
differentiation has resulted in a quantum leap in our
understanding of the fundamental nature of cancer
cells.
While these opportunities exist, the lack of a
common infrastructure has prevented clinical
research institutions from being able to mine and
analyze disparate, multi-level data sources. As a
result, very few cross-site studies and multi-centric
clinical trials are performed and in most cases it isn’t
possible to seamlessly integrate multi-level data.
It is well established that patient recruitment is
often the time-limiting factor for clinical trials. As a
result, clinical trials are gradually turning multi-
centric to limit the time required for their execution
(Sotiriou, 2007).
The ACGT project has been structured within
such a context. It has selected two cancer domains
and has defined three specific trials. These trials
serve a dual purpose. Firstly, they are used for
developing a range of post-genomic analytical
scenarios for feeding the requirement analysis and
elicitation phase of the project, and secondly they
will be used for the validation of the functionality of
the ACGT technologies.
The ACGT trials are in the domain of Breast
Cancer and Wilm’s Tumor (pediatric
nephroblastoma). Specifically:
- The ACGT Test of Principle (TOP) study aims
to identify biological markers associated with
pathological complete response to
anthracycline therapy (epirubicin), one of the
most active drugs used in breast cancer
treatment (Sotiriou, 2003).
- Wilms' tumour, although rare, is the most
common primary renal malignancy in children
and is associated with a number of congenital
anomalies and documented syndromes (Graf,
2007).
In addition to these two clinical trials and on the
basis of data collected for the purpose of their
execution, and in-silico modelling and simulation
experiment is also planned. The aim of this
experiment is to provide clinicians with a decision
support tool able to simulate, within defined
reliability limits, the response of a solid tumour to
therapeutic interventions based on the individual
patient’s multi-level data (Stamatakos, 2007).
2.1 Technical Challenges
ACGT’s vision is to become a pan-European
voluntary network connecting individuals and
institutions and to enable the sharing of data and
tools (see figure 1). In order to achieve its goals and
objectives, ACGT is creating an infrastructure for
cancer research by using a virtual web of trusted and
interconnected organizations and individuals to
leverage the combined strengths of cancer centers
and investigators and enable the sharing of
HEALTHINF 2008 - International Conference on Health Informatics
280

biomedical cancer-related data and research tools in
a way that the common needs of interdisciplinary
research are met and tackled (Tsiknakis, 2006,
Tsiknakis, 2007b).
Considering the current size of clinical trials
(hundreds or thousands of patients) there is a clear
need, both from the viewpoint of the fundamental
research and from that of the treatment of individual
patients, for a data analysis environment that allows
the exploitation of this enormous pool of data
generated.
As a result, a major part of the project is devoted
to research and development in infrastructure
components that are gradually been integrated into a
workable demonstration platform upon which the
selected (and those to be selected during the
lifecycle of the project) clinical studies will be
demonstrated and evaluated against user
requirements defined at the onset of the project.
2.2 Scientific and Functional
Requirements
The real and specific problem that underlies the
ACGT concept is co-ordinated resource sharing and
problem solving in dynamic, multi-institutional, pan-
European virtual organisations. A set of individuals
and/or organisations defined by such sharing
relationships form what we call “an ACGT virtual
organisation (VO)”. Simply stated, the participants
in a multi-centric clinical trial form a VO, which
exists for the duration of a trial or for any other
period of time based on mutual agreements.
The task, therefore, of ACGT is to make data
and tools securely available in this inter-enterprise
environment where and when needed to all
authorised users. As a result, the scientific and
functional requirements for the ACGT platform can
be summarised as follows:
- Virtual Organisation Management: support
for the dynamic creation a VOs, defined as a
group of individuals or institutions who share
the computing and other resources of a "grid"
for a common goal.
- Data federation: seamless navigation across
and access to heterogeneous data sources, both
private and public.
- Data integration: the capacity to pool data
from heterogeneous sources in a scientifically,
semantically and mathematically consistent
manner for further computation.
- Shared services: the development, sharing and
integration of relevant and powerful data
exploitation tools such as tools for
bioinformatics analysis, data mining, modelling
and simulation.
Figure 1: The vision of ACGT. Creating and managing
Virtual Organisations on the Grid who are jointly
participating in the execution of multicentric, post-
genomic Clinical Trials.
The requirements elicitation process that has
taken place in the project, based on input for a
diverse range of users has resulted in the
identification of the following key technical
requirements.
- Flexibility; in other words modularity
(supporting integration of new resources in a
standardised way) and configurability
(accommodating existing and emerging needs).
This is required because (a) The a priori
scientific and functional requirements are broad
and diverse; (b) The data resources to be
federated by the ACGT platform are
characterised by deep heterogeneities in terms
of source, ownership, availability, content,
database design, data organisation, semantics
and so on; and (c) the complexity of the
underlying science, as well as the complexity of
applicable knowledge representation schemas
and applicable scientific algorithms;
- Intuitive access to information; From the
user’s point of view, the ACGT knowledge
management platform must provide relevant
and simple access to information – both in
terms of searching and navigation – and to
services. In addition, it must provide a
dynamically evolving set of validated data
exploration, analysis, simulation, and
modelling services.
- Security; Finally, it must be consistent with the
European ethical and legal framework,
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT OF EFFICIENT KNOWLEDGE DISCOVERY
FROM MULTILEVEL CLINICAL AND GENOMIC DATASETS
281

providing a high degree of trust and security to
its users.
3 THE ACGT INITIAL
ARCHITECTURE
In principle, the requirements for the ACGT
platform can be met by designing a federated
environment articulating independent tools,
components and resources based on open
architectural standards, which is customizable and
capable of dynamic reconfiguration.
In order to fulfill the requirements imposed by
scenarios identified in the ACGT project a
heterogeneous, scalable and flexible environment is
needed and the following technologies, which have
gained momentum in the recent years, have been
considered for adoption:
- Web Services technologies
- Grid technologies
- Semantic web technologies
Although initially separated, these technologies
are currently converging in a complementary way.
Considering that the amount of data generated in
the context of post-genomic clinical trials is
expected to rise to several gigabytes of data per
patient in a close future access to high-performance
computing resources will be unavoidable. Hence,
Grid computing (Foster, 2001) appears as a
promising technology. Access and use of Grid-based
resources is thus an integral part of the design of the
infrastructure.
From the technical point of view, the
requirements identified can be met using a
distributed/federated, multi-layer, service oriented,
and ontology driven architecture. The ACGT
project decided to build on open software
frameworks based on WS-Resource Framework
(WSRF) and Open Grid Service Architecture
(OGSA), the de facto standards in Grid computing.
Building on concepts and technologies from both
the Grid and Web services communities, OGSA
defines uniform exposed service semantics (the Grid
service); defines standard mechanisms for creating,
naming, and discovering transient service instances;
provides location transparency and multiple protocol
bindings for service instances; and supports
integration with underlying native platform
facilities.These standards are implemented in the
middleware selected, namely Globus Toolkit 4 (GT4
- http://www.globus.org/ ).
An overview of the ACGT system layered
architecture is given in Fig. 2, which is shortly
presented in the sequel.
A layered approach has been selected for
providing different levels of abstraction and a
classification of functionality into groups of
homologous software entities (Tsiknakis, 2007a,
Rueping, 2007). In this approach we consider the
security services and components to be pervasive
throughout ACGT so as to provide both for the user
management, access rights management and
enforcement, and trust bindings that are facilitated
by the Grid and domain specific security
requirements like pseudonymization and
anonymization.
Figure 2: The ACGT layered architecture and its main
services.
In specifying the initial architecture of the ACGT
technological platform, architectural specifications
of other relevant projects have been thoroughly
studied. Of particular relevance are the Cancer
Biomedical Informatics Grid (caBIG -
https://cabig.nci.nih.gov/) in the US and the
CancerGrid (http://www.cancergrid.eu/) project in
the UK.
3.1 Heterogeneous Biomedical
Databases Integration
Distributed and heterogeneous databases, created in
the context of mutli-centric post-genomic clinical
trials on cancer, need to be seamlessly accessible
and transparently queried in the context of a user’s
discovery driven analytical tasks. A central
challenge, therefore, to which ACGT needs to
respond, is the issue of semantic integration of
heterogeneous biomedical databases.
HEALTHINF 2008 - International Conference on Health Informatics
282

The process of heterogeneous database
integration may be defined as “the creation of a
single, uniform query interface to data that are
collected and stored in multiple, heterogeneous
databases.” Several varieties of heterogeneous
database integration are useful in biomedicine. The
most important ones are:
Vertical integration. The aggregation of
semantically similar data from multiple
heterogeneous sources. For example, a “virtual
repository” that provides homogeneous access
to clinical data that are stored and managed in
databases across a regional health information
network is reported in (Katehakis, 2007) and
(Lesch, 1997).
Horizontal integration. The composition of
semantically complementary data from multiple
heterogeneous sources. For example, systems
that support complex queries across genomic,
proteomic, and clinical information sources for
molecular biologists are reported in (Stevens,
2000) and (Gupta, 2000).
The approach adopted in ACGT is based on the use
of domain ontologies, acting as the global schema in
a Local-as-View (LAV) integration methodology.
Detailed presentation of the data integration
architecture of the project and the tools and services
utilized for this purpose is outside the scope of this
paper. Such a detailed presentation of the data
integration architecture can be found at (Anguita,
2007).
4 KNOWLEDGE DISCOVERY
SERVICES
Once these multilevel clinical and genomic data are
integrated, they can be mined to extract new
knowledge that can be useful in topics such clinical
diagnosis, therapy, prevention and, of course, the
design of new studies (such as in the case of ACGT,
clinico-genomic trials).
Knowledge discovery in clinico-genomic data
presents a new array of challenges since it differs
significantly from the original problems of data
analysis that prompted the development of Grid
technologies. The exploitation of semantics
information in the description of data sources and
data analysis tools is of high importance for the
effective design and realization of knowledge
discovery processes. Semantics are usually made
concrete by the adoption of metadata descriptions
and relevant vocabularies, classifications, and
ontologies. In ACGT these semantics descriptions
are managed by the Grid infrastructure and therefore
the knowledge discovery services build and operate
on a Knowledge Grid platform (Cannataro, 2003).
4.1 Workflows
The Workflow Management Coalition (WFMC,
http://www.wfmc.org/) defines a workflow as "The
automation of a business process, in whole or part,
during which documents, information or tasks are
passed from one participant to another for action,
according to a set of procedural rules". In other
words a workflow consists of all the steps and the
orchestration of a set of activities that should be
executed in order to deliver an output or achieve a
larger and sophisticated goal. In essence a workflow
can be abstracted as a composite service, i.e. a
service that is composed by other services that are
orchestrated in order to perform some higher level
functionality.
The aim of the ACGT workflow environment is
to assist the users in their scientific research by
supporting the ad hoc composition of different data
access and knowledge extraction and analytical
services into complex workflows. This way the users
can extend and enrich the functionality of the ACGT
system by reusing existing ACGT compliant
services and producing “added value” composite
services. This reuse and composition of services is in
some sense a programming task where the user
actually writes a program to realize a scenario or to
test a scientific hypothesis.
In order to support the ACGT users to build and
design their workflows a visual workflow
programming environment has been designed.
It is a web based workflow editor and designer
that is integrated into the rest of ACGT system so as
to take advantage of the Grid platform and the
ACGT specific infrastructure and services. In
particular, this workflow designer features a user
friendly Graphical User Interface (GUI) that
supports the efficient browsing and searching of the
available ACGT services and their graphical
interconnection and manipulation to construct
complex scientific workflows. The choice of a
graphical representation of the workflow and the
support for ‘point-and-click’ handling of the
workflow graph was made on the basis that this is
more intuitive for the users and increases their
productivity. Additional features that also take
advantage of the metadata descriptions of services
include the validation in the design phase of the
workflows in order to reduce or even eliminate the
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT OF EFFICIENT KNOWLEDGE DISCOVERY
FROM MULTILEVEL CLINICAL AND GENOMIC DATASETS
283

incorrect combination of processing units and the
provision of a “service recommendation”
functionality based on the data types and data
formats of inputs and outputs, and are currently
under development.
Figure 3: The AWESOME (Acgt Workflow Editor
Supporting Online bioMedical invEstigations) Workflow
editor developed in ACGT.
The architecture of the workflow environment
also includes a server side component for the actual
execution (“enactment”) of workflows. Each
workflow is deployed as a “higher order”, composite
service and the Workflow Enactor is the Grid
enabled component responsible for the invocation,
monitoring, and management of running workflows.
The standard workflow description language WS-
BPEL(http://www.oasis-open.org/committees/
tc_home.php?wg_abbrev=wsbpel) has been selected
as the workflow description format and being a
standard it enables the separation of the workflow
designer from the workflow enactor and facilitates
their communication and integration: the designer is
a “rich internet application” running inside the users’
browsers that stores the workflows in WS-BPEL
format into a workflow specific repository whereas
the enactor is an ACGT service running into the
ACGT Grid that “revitalizes” the persisted
workflows as new services.
4.2 Data, Service and Workflow
Metadata
Seamless integration of applications and services
requires substantial meta-information on algorithms
and input/output formats if tools are supposed to
interoperate. Furthermore, assembly of tools into
complex “discovery workflows” will only be
possible if data formats are compatible and semantic
relationships between objects shared or transferred
in workflows are clear. In achieving such
requirements the use of meta-data is important. As a
result, in ACGT we focus on the systematic adoption
of metadata to describe Grid resources, to enhance
and automate service discovery and negotiation,
application composition, information extraction, and
knowledge discovery (Wegener, 2007). Metadata is
used in order to specify the concrete descriptions of
things. These descriptions aim to give details about
the nature, intent, behaviour, etc. of the described
entity but they are also data that can be managed in
the typical ways so this explains the frequently used
definition: “metadata are data about data”.
Examples of this data are: research groups
participating in a CT and publishing the data sets,
data types that are being exposed, analytical tools
that are published, the input data format required by
these tools and the output data produced, and so
forth. Some of the types of metadata that have been
identified are:
- Contact Info: Contact info and other
administrative data about a site participating in
a CT who shares information on the grid.
- Data Type: The data type that a site is exposing
and the context upon which this data was
generated.
- Data Collection Method: This would include the
name of the technique or the platform that was
used to perform the analysis (e.g. Affymetrix),
its model and software version, etc.
- Ontological Category: An ontological category
describes a particular concept that the dataset
exposes or a tool operates upon.
4.3 Analytical Services Metadata
Similarly the identified analytical services’ metadata
descriptions fall into the following categories:
- the task performed by the service; that is the
typology of the analytical data analysis process
(e.g., feature/gene selection, sample/patient
categorisation, survival analysis etc);
- the steps composing the task and the order in
which the steps should be executed;
- the method used to perform an
analytical/bioinformatics task;
- the algorithm implemented by the service;
- the input data on which the service works;
- the kind of output produced by the service;
Our ultimate challenge is to achieve the
implementation of semantically aware Grid services.
In achieving this objective, a service ontology and a
corresponding metadata repository is being
HEALTHINF 2008 - International Conference on Health Informatics
284
developed to provide a single point of reference for
these concepts and to support reasoning of concept
expressions.
5 THE ACGT SECURITY
FRAMEWORK AND ITS
SERVICES
We recognise that the sharing of multilevel data
outside the walls of a hospital or a research
organisation generates complex ethical and legal
issues. It is also well known that the concerns
around “security issues” have been one of the major
obstacles that have inhibited wider adoption of
information technology solutions in the healthcare
domain. As a result we have devoted significant
efforts in the study and analysis of the ethical and
legal issues related to cross-institutional sharing of
post-genomic data sets.
Based on such an approach we concluded that
trust and security must to be addressed at multiple
levels; these include (a) infrastructure, (b)
application access, (c) data protection, (d) access
control, which would be policy-governed, and (e)
privacy-enhancing technology, such as de-
identification.
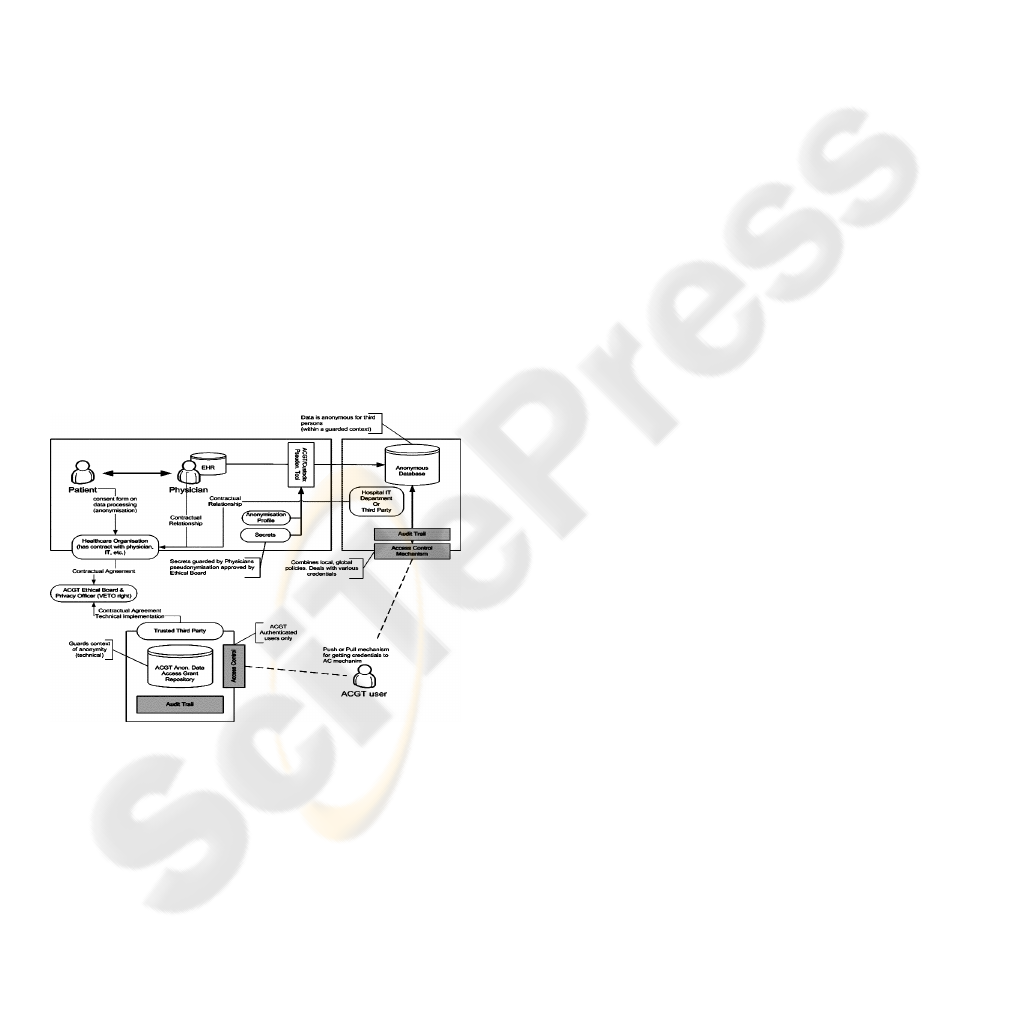
Figure 4: Overview of the ACGT security framework –
actors, procedures and technological services.
The European Directive on Data Protection
(http://www.cdt.org/privacy/eudirective/EU_Directi
ve_.html) deals with the protection of personal data
and imposes many restrictions on its use. In order to
allow ACGT partners to handle and exchange
medical data in conformance with the requirements
of European Directive on Data Protection, an
advanced Data Protection Framework has been
designed. This framework (illustrated on figure 4)
achieves this goal through an integrated approach
that includes technical requirements but also policies
and procedures. Some of the aspects of the Data
Protection framework are (a) Anonymization or
pseudonymisation of the data, (b) a Trusted Third
Party (TTP) pseudonymisation and a corresponding
pseudonymisation tool, (c) technology supported
measures to control the anonymity context, (d) an
ACGT data protection board (acting as a Trusted
Third Party) responsible for issuing credentials for
data access to authorised users, and (e) definition of
the necessary consent forms and legal agreements
that need to be signed by all members of any ACGT
Virtual Organisation.
Description of the technical details of the
security architecture of ACGT (the data protection
framework) goes beyond the scope of the current
article. Nevertheless, the main message that we
want to stress is the fact that a well designed set of
both technological as well as procedural measures
have been taken, so that a high degree of trust and
security is build in the final infrastructure to be
delivered.
6 CREATING AND SHARING
ACGT COMPLIANT SERVICES
Achieving the level of automation, that is
graphically depicted in Figure 3, requires the
creation of highly interoperable services. In turn
creating a service involves describing, in some
conventional manner, the operations that the service
supports; defining the protocol used to invoke those
operations over the Internet; and operating a server
to process incoming requests (Foster, 2005).
Although a fair amount of experience has been
gained with the creation of services and applications
in different science domains, significant problems do
still remain, especially with respect to
interoperability, quality control and performance.
These are issues to which ACGT focuses, and these
are briefly discussed in the next subsections.
6.1 Interoperability and Re-use
Services have little value if others cannot discover,
access, and make sense of them. Yet, as Stein has
observed (Stein, 2002), today’s scientific
communities too often resemble medieval Italy’s
collection of warring city states, each with its own
legal system and dialect. Available technological
(i.e. Web services) mechanisms for describing,
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT OF EFFICIENT KNOWLEDGE DISCOVERY
FROM MULTILEVEL CLINICAL AND GENOMIC DATASETS
285

discovering, accessing, and securing services
provide a common alphabet, but a true lingua franca
requires agreement on protocols, data formats, and
ultimately semantics (d. Roure, 2003). In the ACGT
project we are paying particular attention on these
issues, and especially on the issue of semantics (see
section on metadata).
6.2 Management
In a networked world, any useful service will
become overloaded. Thus, we need to control who
uses services and for what purposes. Particularly
valuable services may become community resources
requiring coordinated management. Grid
architectures and software can play an important role
in this regard and ACGT is focusing on exploiting
these opportunities made available by Grid
computing.
6.3 Quality Control
As the number and variety of services grow and
interdependencies among services increase, it
becomes important to automate previously manual
quality control processes—so that, for example,
users can determine the provenance of a particular
derived data product (Goble, 2004). The ability to
associate metadata with data and services can be
important, as can the ability to determine the identity
of entities that assert metadata, so that consumers
can make their own decisions concerning quality.
7 DISCUSSION AND
CONCLUSIONS
In this paper, we consider a world where biomedical
software modules and data can be detected and
composed to define problem-dependent applications.
We wish to provide an environment allowing
clinical and biomedical researchers to search and
compose bioinformatics and other analytical
software tools for solving biomedical problems. We
focus on semantic modelling of the requirements of
such applications using ontologies.
The project has conceived an overall architecture
for an integrating biomedical sciences platform. The
infrastructure being developed uses a common set of
services and service registrations for the entire
clinical trial on cancer community. We are currently
focusing on the development of the core set of
components up to a stage where they can effectively
support in silico investigation. Initial prototypes
have been useful in crystallizing requirements for
semantics.
The project has set up cross-disciplinary task
forces to propose guidelines concerning issues
related to data sharing, for example legal, regulatory,
ethical and intellectual property, and is developing
enhanced standards for data protection in a web
(grid) services environment.
In addition the project is developing
- standards and models for exposing web
services (semantics), scientific services, and
the properties of data sources, datasets,
scientific objects, and data elements;
- new, domain-specific ontologies, built on
established theoretical foundations and
taking into account current initiatives,
existing standard data representation
models, and reference ontologies;
- innovative and powerful data exploitation
tools, for example multi-scale modelling
and simulation;
- standards for exposing the properties of
local sources in a federated environment;
- a biomedical GRID infrastructure offering
seamless mediation services for sharing
data and data-processing methods and
tools;
- advanced security tools including
anonymisation and pseudonymisation of
personal data according to European legal
and ethical regulations;
- a Master Ontology on Cancer and use
standard clinical and genomic ontologies
and metadata for the semantic integration of
heterogeneous databases;
- an ontology based Trial Builder for helping
to easily set up new clinico-genomic trials,
to collect clinical, research and
administrative data, and to put researchers
in the position to perform cross trial
analysis;
- data-mining services in order to support
and improve complex knowledge discovery
processes;
- an easy to use workflow environment, so
that biomedical researchers can easily
design their “discovery workflows” and
execute them securely on the grid.
A range of demonstrators, stemming from the
user defined scenarios, together with these core set
of components are currently enabling us to both
begin evaluating and gathering additional and more
concrete requirements from our users. These will
HEALTHINF 2008 - International Conference on Health Informatics
286

allow us to improve and refine the facilities of the
ACGT services.
ACKNOWLEDGEMENTS
The authors would like to thank all members of the
ACGT consortium who are actively contributing to
addressing the R&D challenges faced. The ACGT
project (FP6-2005-IST-026996) is partly funded by
the EC and the authors are grateful for this support.
REFERENCES
Anguita, A., et al, 2007. Solving semantic heterogeneities
and integration between clinical and image databases
in post-genomic clinical trials, Proc. of Personalised
Healthcare (phealth2007) Conference (sponsored by
the IEEE Engineering in Medicine
and Biology Society (EMBS)), Chalkidiki, Greece.
Cannataro M. and Talia, D., 2003. KNOWLEDGE Grid -
An Architecture for Distributed Knowledge
Discovery. CACM, vol. 46, no 1, pages 89-93.
d. Roure, D., Jennings N. R. and Shadbolt, N., 2003. The
Semantic Grid: A future e-Science infrastructure, in:
Grid Computing: Making The Global Infrastructure a
Reality. (Eds.) Berman, F., Hey, A. J. G. e Fox, G.,
John Wiley & Sons, pages 437-470.
Foster, I., Kesselman, C., Tuecke, S., 2001. The Anatomy
of the Grid: Enabling Scalable Virtual Organizations.
International Journal of High Performance Computing
Applications, vol. 15, no. 3, pages 200—222.
Foster, I., 2005. Service oriented Science. Science, vol
308, no. 5723, pages 814-817.
Goble, C. , Pettifer, S., Stevens, R., 2004. The Grid:
Blueprint for a New Computing Infrastructure,
Morgan Kaufmann, San Francisco, ed. 2, pages 121–
134.
Graf, N., 2007. The importance of an ontology based
clinical data management system (OCDMS) for
clinico-genomic trials in ACGT (Advancing Clinico-
Genomic Trials on Cancer). In Proc. of the
International Society of Paediatric Oncology
Conference 2007, Mumbai, India (to appear).
Gupta, A., Ludascher, B and Martone, M. E., 2000.
Knowledge-based Integration of Neuroscience Data
Sources. In proc. of the 12th Intl. Conference on
Scientific and Statistical Database Management
(SSDBM), IEEE Computer Society, Berlin.
Katehakis, D.G., et al, 2007. Delivering a Lifelong
Integrated Electronic Health Record based on a
Service Oriented Architecture. IEEE Transactions on
Information Technology in Biomedicine (to appear),
available at:
http://ieeexplore.ieee.org/xpl/tocpreprint.jsp?isnumber
=26793&punumber=4233)
Leisch E., et. al, 1997. A Framework for the Integration of
Distributed Autonomous Healthcare Information
Systems. Medical Informatics, Vol. 22, No. 4, pages.
325-335.
Roses A.D., 2000. Pharmacogenomics and the practice of
medicine. Nature, 405, pages 857-865.
Rüping, S. et al, 2007. Extending workflow management
for knowledge discovery in clinico-genomic data. In
Nicolas Jacq et. al., editor, Proceedings of HealthGrid
2007, volume 126 of Studies in Health Technology
and Informatics, pages 184–193. IOS Press.
Schuppe-Koistinen, I., 2007. The application of metabolic
profiling technologies in biomarker discovery during
drug R&D. In Proc. Pharmaceutical Science World
Congress 2007 (PSWC2007), Amsterdam, The
Netherlands.
Sotiriou, C., et al., 2003. Breast cancer classification and
prognosis based on gene expression profiles from a
population-based study. Proc Natl Acad Sci USA, vol.
100, no. 18, pages 10393-8.
Sotiriou C., Piccart, M.J., 2007. Taking gene-expression
profiling to the clinic: when will molecular signatures
become relevant to patient care?, Nature Reviews,
Vol. 7, July 2007, pages 545-553.
Stamatakos, G.S. et al, 2007. The “Oncosimulator”: a
multilevel, clinically oriented simulation system of
tumor growth and organism response to therapeutic
schemes: towards the clinical evaluation of in silico
oncology. In Proceedings of the 29th Annual
International Conference of the IEEE EMBS, Cité
Internationale, Lyon, France, August 23-26, pages
6628-6631.
Stevens R., et al., 2000. TAMBIS: transparent access to
multiple bioinformatics information sources.
Bioinformatics, vol 16, no. 2, pages. 184–185.
Stein, L., 2002. Creating a Bioinformatics Nation. Nature,
317, pages. 119-120.
Tsiknakis M., et al, 2006. Building a European Biomedical
Grid on Cancer: The ACGT Integrated Project. In
Proc. HealthGrid 2006 Conference, Stud. Health
Technol. Inform., vol.120, pages 247-58.
Tsiknakis M., et al, 2007a. A Semantic Grid Infrastructure
Enabling Integrated Access and Analysis of Multilevel
Biomedical Data in Support of Post-Genomic Clinical
Trials on Cancer, IEEE Transactions on Information
Technology in Biomedicine (to appear), DOI:
10.1109/TITB.2007.903519, available at:
http://ieeexplore.ieee.org/xpl/tocpreprint.jsp?isnumber
=26793&punumber=4233
Tsiknakis M, et al, 2007b. Developing a European Grid
infrastructure for cancer research: vision, architecture,
and services. ecancermedicalsceince Journal, DOI:
10.3332/eCMS.2007.56.
Wegener, D., et. Al.. 2007. GridR: An R-based grid-
enabled tool for data analysis in ACGT clinico-
genomics trials. In proc. of the eScience Conference
2007, Bangalore, India, (accepted, to appear).
A SEMANTIC GRID SERVICES ARCHITECTURE IN SUPPORT OF EFFICIENT KNOWLEDGE DISCOVERY
FROM MULTILEVEL CLINICAL AND GENOMIC DATASETS
287
