
ONTOLOGY BASED INTEGRATION OF DISTRIBUTED AND
HETEROGENEOUS DATA SOURCES IN ACGT
Luis Martín, Alberto Anguita, Víctor Maojo
Biomedical Informatics Group, Artificial Intelligence Laboratory
School of Computer Science, Universidad Politécnica de Madrid, Campus de Montegancedo S/N
28660 Boadilla del Monte, Madrid, Spain
Erwin Bonsma, Anca Bucur, Jeroen Vrijnsen
Phillips Research, Healthcare System Architecture, High Tech Campus 37, 5656 AE Eindhoven, The Netherlands
Mathias Brochhausen, Christian Cocos, Holger Stenzhorn
IFOMIS, Universität des Saarlandes, Postfach 151150, 66041 Saarbrücken, Germany
Manolis Tsiknakis, Martin Doerr, Haridimos Kondylakis
Institute of Computer Science
Foundation for Research and Technology - Hellas, GR-71110 Heraklion, Crete, Greece
Keywords: Ontology-based Biomedical Database Integration, Semantic Mediation, Ontologies, Post-genomic Clinical
Trials, Service Oriented Architectures.
Abstract: In this work, we describe the set of tools comprising the Data Access Infrastructure within Advancing
Clinico-genomic Trials on Cancer (ACGT), a R&D Project funded in part by the European. This
infrastructure aims at improving Post-genomic clinical trials by providing seamless access to integrated
clinical, genetic, and image databases. A data access layer, based on OGSA-DAI, has been developed in
order to cope with syntactic heterogeneities in databases. The semantic problems present in data sources
with different nature are tackled by two core tools, namely the Semantic Mediator and the Master Ontology
on Cancer. The ontology is used as a common framework for semantics, modelling the domain and acting as
giving support to homogenization. SPARQL has been selected as query language for the Data Access
Services and the Mediator. Two experiments have been carried out in order to test the suitability of the
selected approach, integrating clinical and DICOM image databases.
1 INTRODUCTION
Data integration across heterogeneous data sources
and data aggregation across different aspects of the
biomedical spectrum is at the centre of current
biopharmaceutical R&D. A technological
infrastructure supporting such a knowledge
discovery process should, ideally, allow for:
Data to be searched, queried, extracted,
integrated and shared in a scientifically and
semantically consistent manner across
heterogeneous sources, both public and
proprietary, ranging from chemical structures
and omics to clinical trials data;
Discovery and invocation of scientific tools
that are shared by the community, rather than
repeatedly developed by each and every
organisation that needs to analyse their data
and
Both the sharing of tools, and their
integration as modules in a generic
framework, applied to relevant dynamic
301
Martín L., Anguita A., Maojo V., Bonsma E., Bucur A., Vrijnsen J., Brochhausen M., Cocos C., Stenzhorn H., Tsiknakis M., Doerr M. and Kondylakis H.
(2008).
ONTOLOGY BASED INTEGRATION OF DISTRIBUTED AND HETEROGENEOUS DATA SOURCES IN ACGT.
In Proceedings of the First International Conference on Health Informatics, pages 301-306
Copyright
c
SciTePress

datasets. We refer to this process as
“discovery driven scientific workflows” which
ideally would also execute fast and in an
unsupervised manner.
Needless to say that our current inability to
efficiently share data and tools, in a secure and
efficient way, is severely hampering the research
process. The objective of the Advancing Clinico-
Genomic Trials on Cancer (ACGT) project is to
contribute to the resolution of these problems
through the development of a unified technological
infrastructure which will facilitate the seamless and
secure access and analysis, of multi-level clinical
and genomic data enriched with high-performing
knowledge discovery operations and services in
support of multi-centric, postgenomic clinical trials.
Integrated access to heterogeneous biomedical
data is at the core of the problems that need to be
resolved. This paper presents the main
methodological and technological challenges
addressed in the implementation of an ontology-
based data integration architecture within the context
of the ACGT project. Emphasis is given to the
description of the ACGT Data Access Architecture
which is comprised by a set of key services, namely
the ACGT-Data Access Services, the ACGT-
Semantic Mediator, and the ACGT-Master
Ontology, as well as additional dedicated tools.
While the first two services provide the means to
resolve syntactic and semantic heterogeneities when
accessing integrated databases, the latter acts as a
core resource supporting the data integration
process.
2 BACKGROUND
Database Integration aims at facilitating users in
querying sets of heterogeneous sources of
information in an intuitive and transparent way. The
research community has been dealing with different
kinds of methods during the last decade, namely
Data Warehousing (Kimball, 1996), Federated
Database Systems (Sheth, 1990), Mediator-based
approaches (Wiederhold, 1992), and other hybrid
approaches. From the technical point of view, three
categories can be differentiated, namely data
translation, query translation and information
linkage.
In Data Translation, data from the different
databases are integrated in a centralized repository.
Before the integration, these data must be modified
in order to fit the requirements of the unified
schema—the central repository has its own schema
different from the ones belonging to the underlying
databases. The most popular example of a DT-based
technology is Data Warehousing, which is now in its
industrial exploitation phase.
By contrast, Query Translation does not perform
actual integration of data, but transformation of a
query when it is launched. A mediation software
offers a representation of a virtually integrated set of
databases to the users. The user is able to build and
launch a query based on this representation. The
mediator receives the query and transforms it into a
set of dedicated sub-queries for the underlying
databases. After their actual execution in the
corresponding databases, the results are integrated
by the mediator software to be presented to the user.
On the other hand, Information Linkage just defines
cross-reference links between databases to perform
database integration. Some examples of usage of IL
are MEDLINE, GENBANK, OMIM, and the World
Wide Web itself.
There exist two main ways to deal with Query
Translation: Global as View and Local as View. In
both approaches the system has a description of the
domain. In Local as View, views representing the
databases are described using the knowledge
contained in the global schema. In Local as View no
additional work apart from defining a single view is
necessary when a new database needs to be
integrated in the system. However, translation of
queries becomes leads to performance problems
(Abiteboul, 1998) (Ullman, 1997). Conversely, in
Global as View (Cali, 2001) a global model is built
using information from the underlying databases and
from the domain model. Query translation in Global
as View is straightforward, since the links are
actually stored in the schema, but it needs of a global
revision when new sources are added.
During the last years, ontologies have been used
as global domain models in database integration,
obtaining promising results, mainly in the fields of
biomedicine and bioinformatics. Biomedical
ontologies have adopted the role of domain
homogenizing tools in the last decade. We
distinguish three major classes of biomedical
ontologies: Generic Medical Ontologies—dealing
with the entire domain of Medicine—, Specific
Medical Ontologies—describing a single domain
within Medicine—, and Specific Biomedical
Ontologies—supporting a specific biomedical
domain. Some examples of these three categories
are:
Generic Medical Ontologies: SNOMED CT
(SNOMED, 2007), UMLS (Lindberg, 1990).
HEALTHINF 2008 - International Conference on Health Informatics
302

and GALEN (GALEN, 2007), HL7 RIM
(HL7, 2007). Both SNOMED CT and UMLS
have been proved to be theoretically unsound
(Ceusters, 2003). HL7 RIM, even though
widely used, has been subjected to a number
of criticisms that also question its theoretical
soundness (Smith, 2006).
Specific Medical Ontologies: The
Foundational Model of Anatomy (FMA,
2007) is a highly stabile and rigorously
developed ontology. One system frequently
mentioned when talking about the state of the
art in ontology-based cancer research and
management is caCORE (caCORE, 2007), a
highly developed environment making use of
UMLS and NCI Thesaurus and Metathesaurus
representations (NCI, 2007).
Specific biomedical Ontologies: Gene
Ontology (GO, 2007) is one example. Other
examples can be found at the OBO Foundry
(OBO, 2007). There is a high number of
Specific Biomedical Ontologies, and they
follow a variety of different standards. This
shows the importance of quality assessment in
ontology development within this domain.
The following section describes in detail the
database access architecture adopted to integrate
clinical trials databases including image information.
3 THE ACGT DATA ACCESS
INFRASTRUCTURE
The ACGT platform is comprised by a set of
services and resources supporting the different needs
of clinicians and researchers involved in a post-
genomic clinical trial. The ACGT platform
architecture follows a layer based design, as can be
seen in Figure 1.
Figure 1: The ACGT platform architecture.
The ACGT data access infrastructure forms part of
this architecture. This infrastructure is comprised by
three core resources, together with other satellite
tools that give support to the complete data access
task. These core resources are, namely: the ACGT
Master Ontology on Cancer (ACGT-MO), the
ACGT Data Access Services (ACGT-DAS) and the
ACGT Semantic Mediator (ACTG-SM). Figure 2
shows the architecture of the ACGT Data Access
Infrastructure.
Figure 2: The ACGT data access infrastructure.
The next sections give a detailed description of these
three components.
3.1 ACGT-MO
ACGT deals with the integration of data from a
variety of heterogeneous sources. There exists a lack
of standardization among data from different clinical
trials, which leads to a loss in the possible
knowledge exchanging power. Ontology based data
management becomes then a major advantage in the
way to achieve consistency in data collection and
processing policies.
The ACGT-MO employs the resources of a Top
Level Ontology, called Basic Formal Ontology
(BFO, 2007). This choice is based on its proven high
applicability to the biomedical field (Grenon, 2004).
The ACGT Master Ontology inherits BFO’s
foundational principles:
realism
perspectivalism
fallibilism
adequatism
Figure 3 shows the BFO structure.
ONTOLOGY BASED INTEGRATION OF DISTRIBUTED AND HETEROGENEOUS DATA SOURCES IN ACGT
303
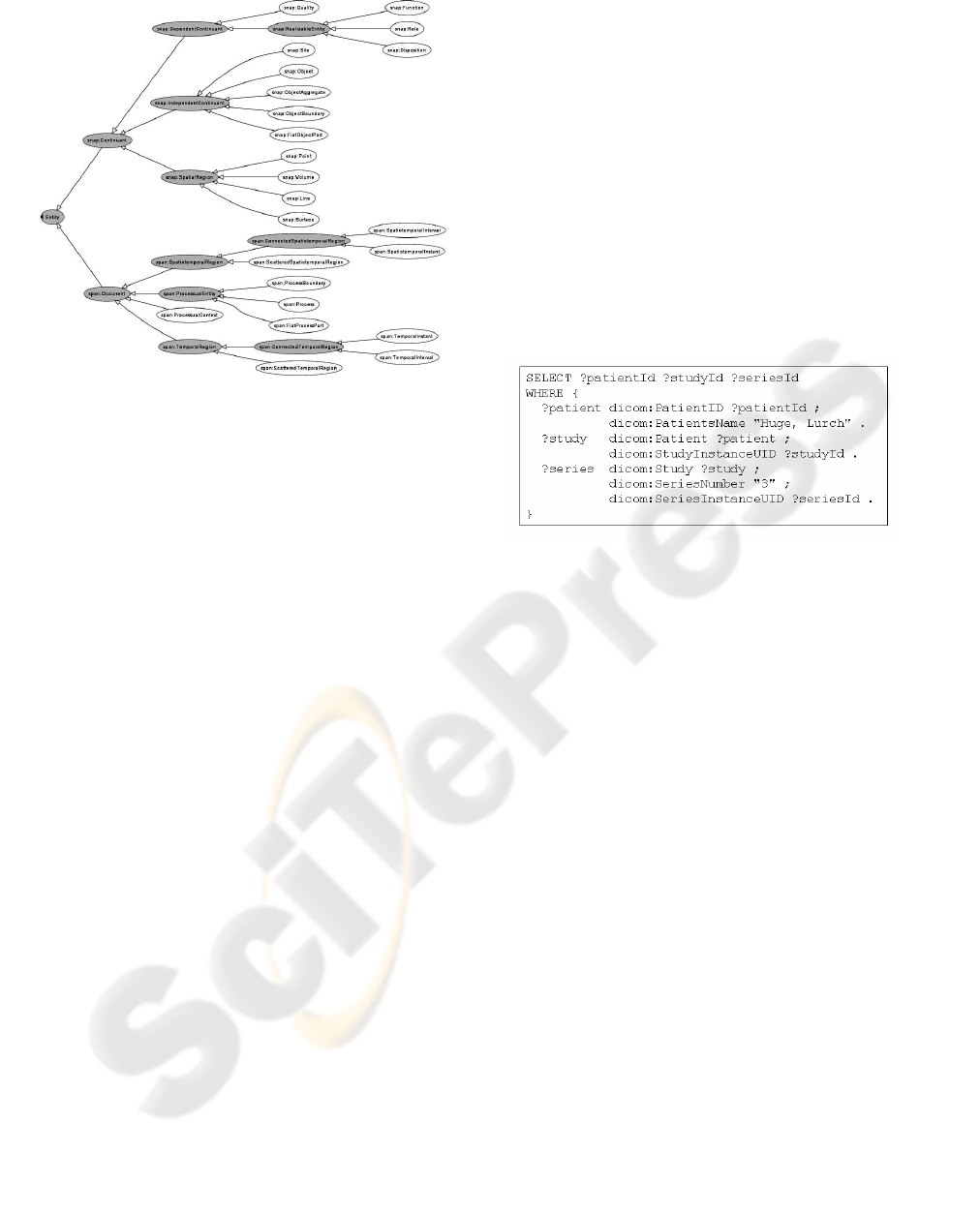
Figure 3: The Basic Formal Ontology.
The ACGT-MO has been developed using the
OWL-DL language, achieving the maximum level of
expressivity to describe the domain of post-genomic
clinical trials on cancer. For its development and
mantainance, the Protégé editor (Protégé, 2007) has
been used.
The ACGT-MO basically contains two sets of
elements, namely i) Classes and ii) Properties. The
former group contains the concepts of the ontology
(the so-called universals) structured in a taxonomy
using is_a type relations to establish links between
classes—e.g., CanonicalBodySubstance is_a
BodySubstance. The latter represents the set of
relations connecting the classes of the taxonomy. In
order to fit the requirements of data integration in
biomedical reality, and to express the truths of
Medicine and Biology, a wide variety of relations
(besides from mere is_a) has been included. A few
examples of structure in the tree of relations are
hasBloodPressure is a child of hasPressure which, in
turn, is a child of hasMagnitude, or hasFunction is a
child of implements. An important part of the
relations list has been imported from the Open
Source Relation Ontology (RO) (RO, 2007).
3.2 ACGT-DAS
The ACGT-DAS provide a means to solve syntactic
heterogeneities—i.e. they provide uniform data
access interface. ACGT-DAS are required also to
export the data schema of each individual source, in
order to aid the clients in building queries.
The ACGT-DAS offer a web service interface.
They have been implemented using the Open Grid
Services Architecture Data Access and Integration
(OGSA-DAI) services (Antonioletti, 2005)
SPARQL (SPARQL, 2007) has been chosen as
the query language. More expressive than its
predecessor RDQL, the language used by an early
version of the mediator, SPARQL offers new
features, becoming an intermediate level (in terms of
expression) language, appropriate for being used as
common query language. It is less expressive than
Structured Query Language (SQL), due to the lack
of support of any form of aggregation. SQL is a
relational specific language, so it cannot be used as
common language by the ACGT-DAS (mainly
because of the selected query translation approach).
On the other hand, it is more expressive than
DICOM.
Figure 4: Example of DICOM query in SPARQL.
A relational database and a DICOM wrapper
have been developed so far. We used D2RQMap
(Bizer, 2004) for the implementation of the
relational databases wrapper. This was a
straightforward process, due to the technology
choices. The development of the wrapper for
querying DICOM image databases was not as direct.
DICOM uses a four-level hierarchical information
model, not a classical relational model. This
structure caused difficulties in the query
transformation process, since SPARQL is more
expressive than DICOM, so the final queries may
not be able to represent the view that is expressed in
the original one. Figure 4 shows an example of a
DICOM query expressed in SPARQL. A set of
special functionalities had to be implemented to
support retrieving of DICOM images as well.
3.3 ACGT-SM
The ACGT-SM aims at solving the semantic
heterogeneities present in databases to support
interoperability and integration. The ACGT-SM is
supported by a set of satellite tools, like the mapping
tool and the data cleaning module among others.
The approach selected to perform database
integration has been Local as View. This decision is
based on the nature of data in the biomedical
HEALTHINF 2008 - International Conference on Health Informatics
304

domain, and more concretely in post-genomic
clinical trials. Local as View based techniques
require less amount of effort when the structure of
data sources changes or when new ones need to be
integrated in the system. However, as stated before,
some performance issues are associated to this kind
of approaches. In order to overcome these
difficulties, the domain representing the integrated
set of databases is constrained. This restriction is
based on requirements specified by the end users.
Semantic heterogeneities are tackled following
an ontology based approach. The ACGT-MO acts as
a semantic framework supporting homogenization.
In Local as View, the ACGT-MO acts as global
schema. The goal of this global schema is twofold:
1) provides a means to build the local views of the
underlying databases, and 2) represents the set of
queries that can be formulated by the users.
SPARQL has been selected as query language
for the ACGT-SM. As said previously, SPARQL is
used as query language by the ACGT-DAS. This
homogeneity allows saving time and memory
resources. When a query is launched through the
ACGT-SM, the software divides it into a set of
dedicated queries for the underlying data access
services, wrapping the actual databases. No interface
is needed to translate these queries, given that the
same query language is used by the ACGT-SM and
the ACGT-DAS.
The results of a query are returned by the
wrappers in XML SPARQL Result format. The
ACGT-SM builds an integrated set of results as an
ontology instance file. These instances are
represented using the OWL ontology description
language. Other formats, such as CSV (Comma
Separated Variables) are supported to fit the
requirements of the Data Analysis Tools in ACGT.
Parallel to the ACGT-SM, an API for creating
mappings between the ACGT-MO and RDF
Schemas of data sources to be integrated has been
developed. This API offers a flexible and generic
approach for creating mappings, and is based on
path mapping. Paths from the ACGT-MO are
mapped to paths from an RDF Schema, providing a
way to translate queries to the ACGT-MO—which
are basically sets of paths—into queries to the RDF
Schema. A graphical interface on top of this API is
being developed as well.
3 EXPERIMENTS AND RESULTS
We have tested the first version of our tools in a case
study including a set of three different sources,
including two clinical relational databases—SIOP
nephroblastoma database and TOP breast cancer
database— and a DICOM repository of images. We
carried out two experiments integrating DICOM
source and each one of the clinical trials databases.
Our tool integrated the sources successfully, and the
generated schemas were validated by experts in the
domain.
The experiments were performed using a
dedicated web interface. This interface was built for
demonstration purposes within the ACGT project,
but it is not the final query interface—this interface
is now in its design phase, and is going to be able to
support more complex queries. Figure 5 shows the
result of the execution of a query combining SIOP
and DICOM data.
Figure 5: Instances retrieved: SIOP and DICOM
integration.
As can be seen in Figure 5, the user can request
the DICOM studies related to the patient selected by
clicking on the relation button.
The experiments showed the suitability of the
approach adopted to cope with data access and
integration in the post-genomic clinical trials
domain. This prototype was developed using Java
and HTML languages.
4 CONCLUSIONS
In this work, we present a set of tools to provide data
access, allowing seamless access and integration of
heterogeneous databases. To this end, a clinical trials
on cancer domain ontology has been developed, the
ACGT-MO, and two core services to overcome
syntactic and semantic heterogeneities, namely the
ACGT-DAS and the ACGT-SM.
The ACGT-MO covers the domain of clinical
trials on cancer, and has been built using Clinical
Report Forms from SIOP and TOP trials. The
ONTOLOGY BASED INTEGRATION OF DISTRIBUTED AND HETEROGENEOUS DATA SOURCES IN ACGT
305

ACGT-MO follows the recommendations of the
OBO Foundry.
The ACGT-DAS resolve syntactic
heterogeneities present in disparate sources of
information. They provide a homogenous query
language, SPARQL, and a web service interface
developed using the OGSA-DAI middleware.
The ACGT-SM is able to process user queries
formulated by means of a global model—i.e. the
ACGT-MO—, and to retrieve information from a set
of integrated heterogeneous databases. The ACGT-
SM is supported by a set of satellite tools tackling
with problems such as mapping and instance
homogenization.
The results obtained in the carried out
experiments prove that this approach can properly
integrate relational and image databases.
In the second phase of the project, we plan to
add new types of sources, such as public web
databases, different file formats—e.g. plain text,
Excel spreadsheets, XML, etc— and microarray
data.
ACKNOWLEDGEMENTS
The authors would like to thank all members of the
ACGT consortium who are actively contributing to
addressing the R&D challenges faced. The ACGT
project (FP6-2005-IST-026996) is partly funded by
the EC and the authors are grateful for this support.
REFERENCES
Abiteboul S., Duschka O., 1998. Complexity of answering
queries using materialized views. In Proc. of the 17th
ACM SIGACT SIGMOD SIGART Symp. on Principles
of Database Systems (PODS’98), pages 254-265.
Antonioletti, M., et al. 2005. The Design and
Implementation of Grid Database Services in OGSA-
DAI- In: Concurrency and Computation: Practice and
Experience, Volume 17, Issue 2-4, Pages 357-376.
BFO, The Basic Formal Ontology. Available at:
http://www.ifomis.uni-saarland.de/bfo [13 oct 2007]
Bizer, C., Seaborne, A. 2004. D2RQ – Treating Non-RDF
Databases as Virtual RDF Graphs. In: Proc. of the 3rd
International Semantic Web Conference (ISWC2004),
Hiroshima, Japan. Poster presentation.
caCORE: A Common Framework for Cancer Data
Management. Available at:
http://ncicb.nci.nih.gov/infrastructure/cacore_overvie
w [13 oct 2007]
Cali, A., De Giacomo, G., Lenzerini, M., 2001. Models for
information integration: Turning local-as-view into
global-as-view. In: Proc. of Int. Workshop on
Foundations of Models for Information Integration
(10th Workshop in the series Foundations of Models
and Languages for Data and Objects).
Ceusters W, Smith B, Kumar A, Dhaen C. 2003. Mistakes
in Medical Ontologies: Where Do They Come From
and How Can They Be Detected?. in: Pisanelli DM
(ed.): Ontologies in Medicine: Proceedings of the
Workshop on Medical Ontologies, Rome, October
2003. IOS Press, Amsterdam.
FMA, The Foundational Model of Anatomy. Available at:
http://fme.biostr.washington.edu:8089/FME/index.htm
l [13 oct 2007]
GALEN, Common Reference Model. Available at:
http://www.opengalen.org [13 oct 2007]
GO, Gene Ontology project. Available at:
http://www.geneontology.org [13 oct 2007]
Grenon, P., Smith, B., Goldberg, L., “Biodynamic
Ontology. 2004. Applying BFO in the Biomedical
Domain,” in: Ontologies in Medicine, D. M. Pisanelli,
Ed., Amsterdam: IOS Press, pp. 20-38.
HL7, Reference Information Model. Available at:
http://www.hl7.org/Library/data-
model/RIM/modelpage_mem.htm. [13 oct 2007]
Kimball R., 1996. The Data Warehouse Toolkit: Practical
Techniques for Building Dimensional Data
Warehouses, John Wiley.
Lindberg C. The Unified Medical Language System
(UMLS) of the National Library of Medicine. Journal
of the American Medical Record Association 1990;
61(5):40-2.
NCI Enterprise Vocabulary Services. Available at:
http://www.nci.nih.gov/cancerinfo/terminologyresourc
es [13 oct 2007]
OBO Foundry ontologies. Available at:
http://obofoundry.org [13 oct 2007]
Protégé, The Ontology Editor and Knowledge Acquisition
System. Available at: http://protege.stanford.edu [13
oct 2007]
RO, OBO Relation Ontology. Available at:
http://www.obofoundry.org/ro [13 oct 2007]
Sheth A. P., Larson J. A., 1990. Federated Database
Systems for Managing Distributed, Heterogeneous,
and Autonomous Databases, ACM Computing
Surveys, 22(3): pp.183-236.
Smith, B, Ceusters, W. HL7 RIM: An Incoherent
Standard, Stud Health Technol. Inform. 2006; 124:
133–38
SNOMED Clinical Terms Core Content. Available at:
http://www.snomed.org/snomedct/coreterms.html [13
oct 2007]
SPARQL Query Language for RDF. Available at:
http://www.w3.org/TR/rdf-sparql-query/ [13 oct 2007]
Ullman J.D., 1997. Information integration using logical
views. In Proc. of the 6th Int. Conf. on Database
Theory (ICDT’97), volume 1186 of Lecture Notes in
Computer Science, pages 19-40. Springer-Verlag.
Wiederhold G., 1992. Mediators in the Architecture of
Future Information Systems, IEEE Computer, 25(3):
pp. 38-49.
HEALTHINF 2008 - International Conference on Health Informatics
306
