
ENDOCARDIAL SEGMENTATION IN CONTRAST
ECHOCARDIOGRAPHY VIDEO WITH DENSITY BASED
SPATIO-TEMPORAL CLUSTERING
Prashant Bansod, U. B. Desai
SPANN Lab., Electrical Engg. Department, Indian Institute of Technology, Bombay, India
Nitin Burkule
Cardiologist, Asian Heart Hospital and Research Center, Mumbai, India
Keywords:
Contrast echocardiography, Left ventricle, Segmentation, Spatio-temporal clustering.
Abstract:
We present a spatio-temporal clustering algorithm for detection of endocardial contours in short axis (SAX)
contrast echocardiographic image sequences. A semiautomatic method for segmentation of left ventricle in
SAX videos is proposed which uses this algorithm and at the same time requires minimal expert intervention.
Expert is required to specify a few candidate points belonging to the contour, only in the first frame of the
sequence. The initial contour is approximated by fitting an ellipse in the region defined by the points speci-
fied. This region is identified as the principal cluster corresponding to the left ventriclular cavity. Later the
density based clustering was applied for regularization on the inital contour. We have extended the DBSCAN
algorithm for identification of the principal cluster corresponding to the left ventricle from the image. The al-
gorithm also incorporates the temporal information from the adjacent frames during the segmentation process.
The algorithm developed was applied to 10 data sets over full cardiac cycle and the results were validated by
comparing computer generated boundaries to those manually outlined by one expert. The maximum error in
the contours detected was ±2.9mm. The spatio-temporal clustering algorithm proposed in this paper offers an
efficient semiautomatic segmentation of heart chambers in 2D contrast echocardiography sequences.
1 INTRODUCTION
Amongst the various medical imaging modalities, two
dimensional (2D) echocardiography is valuable for
patients with heart diseases. It is noninvasive, real
time, easy to use in clinical environment and of-
fers relatively low cost solution as compared to other
modalities (Bridal et. al, 2003). However, for eval-
uation of cardiac functional parameters, segmenta-
tion is to be carried out. Manual segmentation as
routinely carried by experts is time consuming and
tedious due to large image data in different stan-
dard echo views over a full cardiac cycle. Again
the manual method also suffers from inter-observer
and intra-observer variability in measurements (Maes
et. al, 1993). Many researchers have shown image
processing applications to enhance clinical utility of
echocardiography by automated and semiautomated
endocardial border delineation and for evaluation of
functional cardiac parameters (Noble and Boukerroui,
2006). In fact there is a continuous growing de-
mand for the automated segmentation and quantifi-
cation to support professionals in diagnosis. In re-
cent years automated segmentation of heart cham-
bers and in particular the left ventricle has received
significant attention in 2D and 3D echocardiograms.
However automatic edge definition and subsequent
segmentation in echocardiograhic images is difficult
due to presence of speckle noise, poor contrast, inher-
ent dropouts, inter-cavity structures and variability of
data along with orientation and positioning of trans-
ducer (Setaredhan and Soragham, 1996).
In recent years numerous clinical studies have
shown the clinical utility of myocardial contrast
echocardiography (MCE) in quantification of my-
ocardial perfusion, left ventricle (LV) volumes, LV
contours and cardiac functional parameters (Cohen
et.al., 1998). There have been few reports of research
attempts towards the semiautomatic and fully auto-
matic segmentation of left ventricle from 2D con-
trast enhanced echo images (Wolfer et. al, 1999).
A very rigorous work for the segmentation problem
204
Bansod P., B. Desai U. and Burkule N. (2008).
ENDOCARDIAL SEGMENTATION IN CONTRAST ECHOCARDIOGRAPHY VIDEO WITH DENSITY BASED SPATIO-TEMPORAL CLUSTERING.
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 204-209
DOI: 10.5220/0001064102040209
Copyright
c
SciTePress

in low mechanical-index contrast echocardiography
is reported (Zwirn et. al, 2006). It has been shown
that the use of temporal continuity results in better
segmentation as it follows the approach of human ex-
pert in delineation (Mullet-Parada and Noble, 1998).
Typically the dropouts present in the image can be re-
covered by the use of boundary information from the
neighboring frames (Choy et. al, 1998). Researchers
have reported active contour approach (Morales et.
al, 2002), trained deformable models (Garcia et. al,
2003) and active shape model (Pickard et. al, 2004).
Many of the proposed methods have shown results
comparable to expert delineation for good quality im-
ages (Mishra et. al, 2003). However none of the
methods has a generalized applicability for fully auto-
matic or semiautomatic segmentation for the images
acquired in routine clinical environment.
Few researchers have extended the application of
well established data clustering approaches in the
field of medical image segmentation (Celebi et. al,
2005). In this work we have extended the Density-
based Clustering (DBSCAN) approach by including
temporal data and applied for the segmentation of
contrast echo sequences. Our spatio-temporal cluster-
ing algorithm has shown good results in the segmen-
tation of endocardial borders in frames of a sequence
by accommodating temporal information. The user
intervention is minimal and is of the form of specify-
ing five or more candidate points for contour on the
first frame of the sequence.
The paper is organized as follows: In section II
we discuss the density based clustering and its ex-
tension in spatio-temporal clustering technique. In
section III, we present the application of the algo-
rithm for segmentation of endocardial border after fit-
ting the ellipse in the first frame through the points
specified by the user and then to subsequent frames
in the sequence. The contours thus obtained are post
processed and smoothened to obtain final endocardial
borders. In section IV we present the results of the
proposed algorithm and, finally conclusions drawn
and future work is discussed in Section VI.
2 CLUSTERING
Clustering is an important technique in data min-
ing for finding data distributions and patterns in the
underlying one or more dimensional data (Jain and
Dubes, 1988). It has been a active field of research
since last two decades and many novel approaches
have been reported in the literature (Jain et. al,
1999). Clustering has number of upcoming appli-
cation fields, such as statistical data analysis, pat-
tern recognition, image processing, segmentation and
many others. It is the task of grouping similar objects
together with respect to a distance , connectivity, con-
tinuity, relative density in the space or other similarity
measure.
In formal mathematical definition cluster is de-
fined as (Fung, 2001): Let X ∈ R
m×n
be a set of data
items representing a set of m points in x
i
in R
n
. The
goal is to partition X into K groups C
k
such that ev-
ery data that belongs to the same group are more alike
than data in different groups. Each of the K groups is
called a cluster. The result of the algorithm is an injec-
tive mapping X 7−→ C of data items X
i
to clusters C
k
.
The number K might be preassigned by the user or it
can be unknown determined by the algorithm . There
are many approaches to data clustering that vary in
their complexity and effectiveness. For our applica-
tion we have focussed our attention on a single cluster
(K = 1), pertaining to the heart chamber specifically,
the left ventricle in the contrast echcocardiographic
view. In our work we call it as principal cluster. The
assumption of defining only one principal cluster is
valid because the spatial coordinates of the boundary
objects of this principal cluster reflect the endocardial
contour.
2.1 Dbscan for Principal Cluster
Density-based algorithms typically regard clusters as
dense regions of objects in the data space separated by
regions of low density. Thus the main objective lies
in finding regions of high and low densities (Bradley
and Fayyad, 1998). This approach is also capable of
finding arbitrarily shaped clusters in the data space.
Another advantage of these algorithms is that they
are independent of the prior knowledge of the number
of clusters. Hence these are very useful in situations
very clustering can be confined to only in the region
of interest (Han and Kamber, 1998). In contrast en-
hanced short axis echo sequence, the chamber cavi-
ties are filled with micro-bubbles which contribute in
achieving their opacity. This results in a bright re-
gions corresponding to the blood filled areas in an
echo image (Fedele et. al, 1998). In the SAX images
of the left ventricle (LV), a single bright region in the
center of the acoustic window corresponds to the LV
cavity. We treat this central bright region as a single
cluster of interest. As stated earlier it is termed as the
principal cluster for this application. The two global
parameters of density based clustering algorithms are:
• E ps: Maximum radius of the neighborhood.
• MinPts: Minimum number of points in the Eps
neighborhood of a point.
ENDOCARDIAL SEGMENTATION IN CONTRAST ECHOCARDIOGRAPHY VIDEO WITH DENSITY BASED
SPATIO-TEMPORAL CLUSTERING
205
The ellipse fitted through the expert specified points
in the first frames is taken as the starting point for
the density based cluster algorithm. The maximum
radius of the neighborhood E ps is chosen as half of
the major axis of the ellipse. The parameter MinPts
was chosen to be 100 after a study of end systole im-
ages in 44 patients. The core point of the principal
cluster is chosen as the center point of the ellipse fit-
ted through the points specified by the expert in the
first frame. We have chosen four local parameters for
grouping the objects (pixels) in a cluster. These pa-
rameters include the features of the objects like pixel
intensity, gradient threshold, gradient angle and the
angular gradient with respect to the center point of the
region. For cavity boundary, only negative intensity
changes are identified along radial lines from center
point. Again the threshold for gradient (G
T
) was ob-
tained automatically from the histogram statistics and
the coordinates and intensities of the pixels specified
by the expert. The algorithm for our application is
framed as:
• The center of the ellipse is taken as core point.
• The maximum radius is assigned the value of semi
major axis in the first frame.
• Gradient threshold is obtained by histogram of the
frame.
• Density reachable points around the core point
are identified.
• Above steps are repeated for all the frames in the
sequence.
• Border objects of the cluster are determined.
2.2 Spatio-temporal Dbscan
An image is a 2-dimensional(2D) array of pixels de-
fined on a W × H rectangular lattice S = [ (x, y) :
1 ≤ x ≤ W, 1 ≤ y ≤ H ], and is indexed by the co-
ordinate (x, y). Each pixel in a given frame can be
represented by a feature vector. In a video stream, im-
age frames are continuous along the time axis. Thus
a video sequence can be expressed in spatio-temporal
domain. Temporal dimension can be incorporated in
many ways. One of the way is separating the frames
of the sequence with respect to discrete time and to
stack consecutively. We follow this approach in our
application. In a video sequence, the frame to frame
variation in shape and dimension of a given object
depends upon its deformity and forces acting on it.
Hence it is possible to recover a missing segment or
to correct any outlier in the contour from the adjacent
frames if the frame to frame variation is not signifi-
cant. The outliers are detected with the radius of cur-
vature of the extracted contour and the corresponding
points in adjacent frames. We propose temporal con-
tinuity in the neighborhood of three frames:
1. For i
th
frame the j
th
border pixel will lie in the
bounds setup by (i − 1)
th
and (i + 1)
th
frame.
2. Presence of drop out pixels in a frame was taken
by temporal continuity from adjacent frames over
an interval of three fames.
We have used parameter λ as the correction factor
which governs the closeness of the corrected seg-
ment with the temporal frames. Correction is incor-
porated at 360 equidistant points on the contour i.e
j = [1, 2, .....360]. The clustering is recursively car-
ried out to regroup the cluster on the basis of modified
distance and density parameters. These parameters in
turn are function of λ during recursive calls of DB-
SCAN.
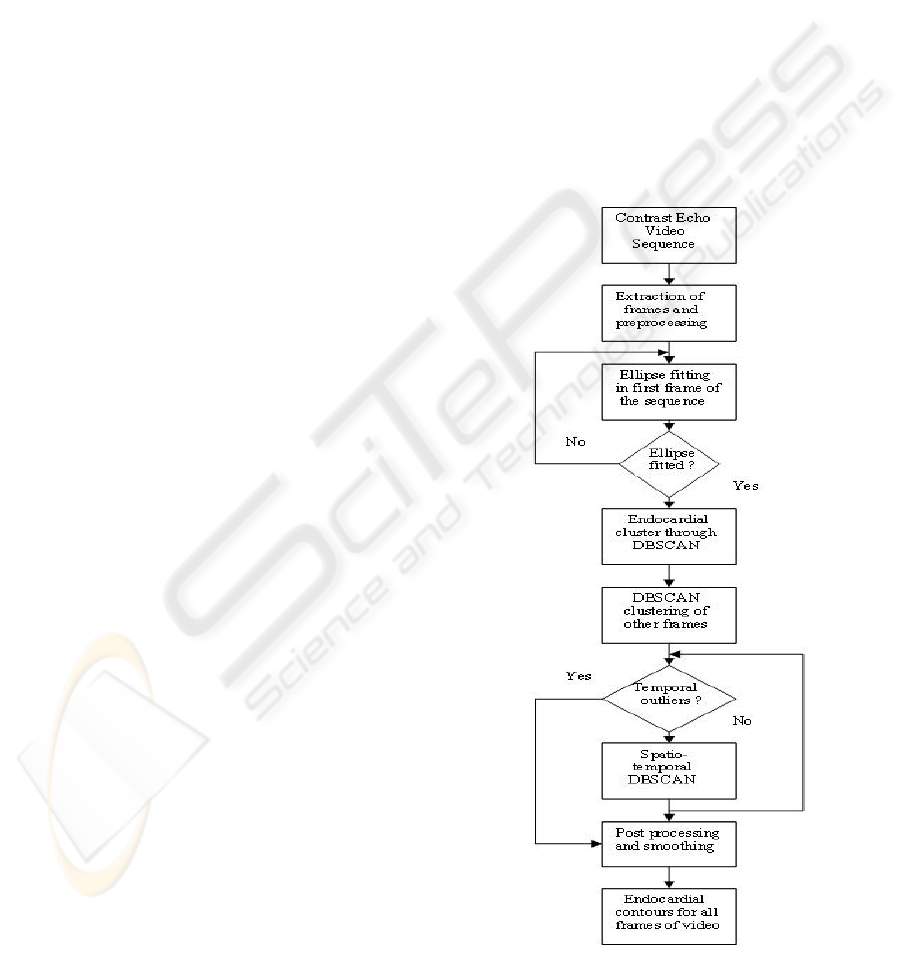
Figure 1: Flow chart for segmentation procedure.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
206
3 SEGMENTATION WITH
T-DBSCAN
In this work, we have applied the spatio-temporal
clustering algorithm to segment the endocardial bor-
der in the contrast enhanced echocardiography videos
of 10 patients. Figure 1. shows the flowchart of the
segmentation procedure. After preprocessing of the
image frames, user specifies candidate boundary pix-
els of the left ventricle in the first frame of the first
frame of the sequence by mouse. The ellipse is fitted
through these points and its parameters are stored for
subsequent processing.
3.1 Image Processing
The echocardiographic videos used in this study were
contrast enhanced short axis apical images at various
levels of LV. These were obtained from different sub-
jects for two to four cardiac cycles. The videos were
acquired on GE Vingmed Ultrasound, VIVID7 in hos-
pital environment under expert guidance. The frames
in each video were 434 x 636 true color with 8 bit bit-
depth in DICOM format. Gray scale conversion with
256 levels was done. The video sequences for one
complete cardiac cycle were used for estimation of
LV border. Echo images contain speckle noise which
lead to incorrect gradient estimation. Hence speckle
reducing anisotropic diffusion (SRAD) filtering was
used (Yongjian and Scott, 2002). They have sug-
gested edge sensitive diffusion for reducing speckles.
In the numerical implementation we used δt = 0.008
and threshold of 5. This reduced the speckles and at
the same time preserved the edge information for fur-
ther feature extraction.
3.2 Elliptical Boundary Approximation
The Initial boundary approximation is carried out in
the first frame of the sequence by fitting a ellipse
through the points specified by the expert. The best
fit ellipse through the points specified is done using
Least Squares Criterion (Fitzgibbon et. al, 1999) A
minimum of five points are to be specified by the ex-
pert, which strongly belong to the endocardial bor-
der for that particular frame. This is the only user
intervention which is required in our scheme. The
standard impixel function of MATLAB is used which
gives the spatial coordinates of the selected points
along with their intensities. The intensities returned
by the function were used in the subsequent proce-
dure for the search.
The generalized CONIC equation of the Ellipse is
given by:
ax
2
+ by
2
+ cx +dy + exy + f = 0 (1)
with a, b and c not all zero and b
2
< 4ac, where all
of the coefficients are real. Again, more than one so-
lution, defining a pair of points (x, y) on the ellipse,
exists. It can be expressed in matrix notation as;
X
T
AX = 0 (2)
where X and A are given by
X = [1 x y]
0
(3)
A =
f 0.5c 0.5d
0.5c a 0.5e
0.5d 0.5e b
(4)
The coordinates of the N chosen points (N >= 5) as
marked by the expert and the equations (2-4) are used
for the determination of the parameter matrix of the
conic representation. The orientation and tilt of the el-
lipse is sought by coefficients in the equation (1) and
incorporated in the evaluation of final ellipse parame-
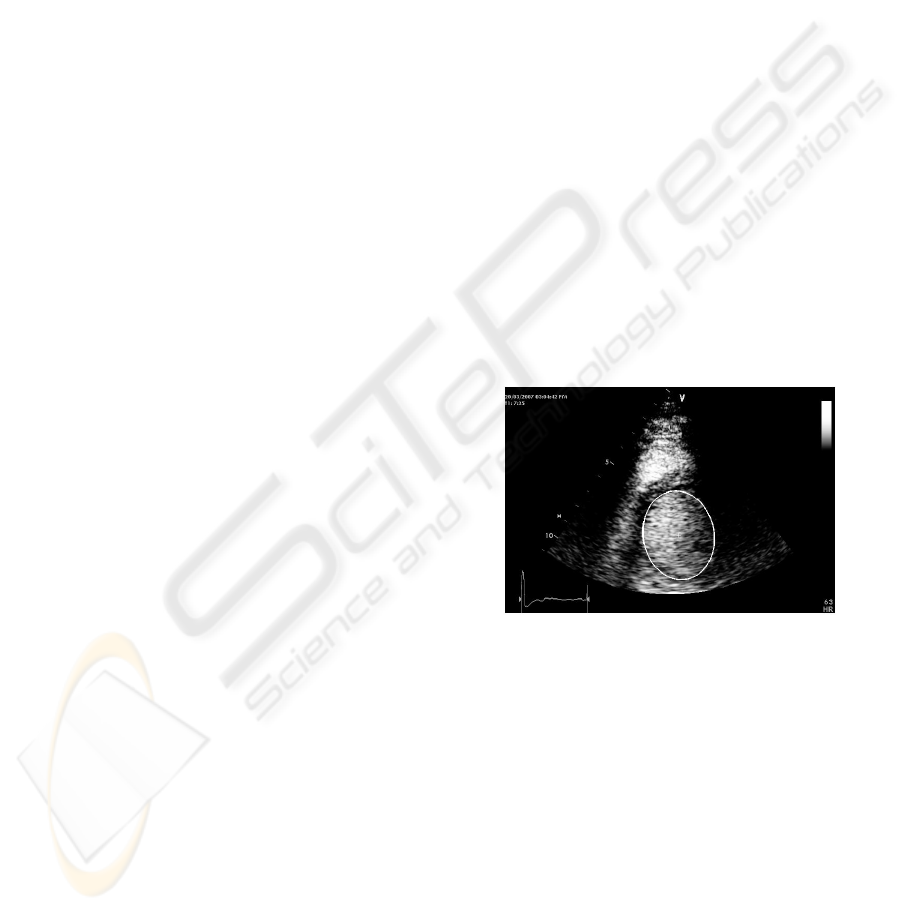
ters using square completion method. Figure 2 shows
the first frame of SAX apical sequence with the de-
tected ellipse and its center point.
Figure 2: Ellipse Fitted in the first frame of the sequence.
3.3 Spatio-temporal Clustering
The DBSCAN clustering algorithms is recursively
called for incorporating corrections in the outliers
with the parameter λ. In our implementation we have
used λ = 0.5 distance units, which gave optimum re-
sults.
3.4 Post Processing and Smoothing
The contour thus obtained was smoothed out by lo-
cally weighted scatter plot smoothing using least
squares linear polynomial fitting. A span of
10 percent was used to implement this standard
ENDOCARDIAL SEGMENTATION IN CONTRAST ECHOCARDIOGRAPHY VIDEO WITH DENSITY BASED
SPATIO-TEMPORAL CLUSTERING
207
MATLAB function. Further smoothing was carried
out by fitting spline through the data points with min-
imizing the maximum square distance between the
data points.
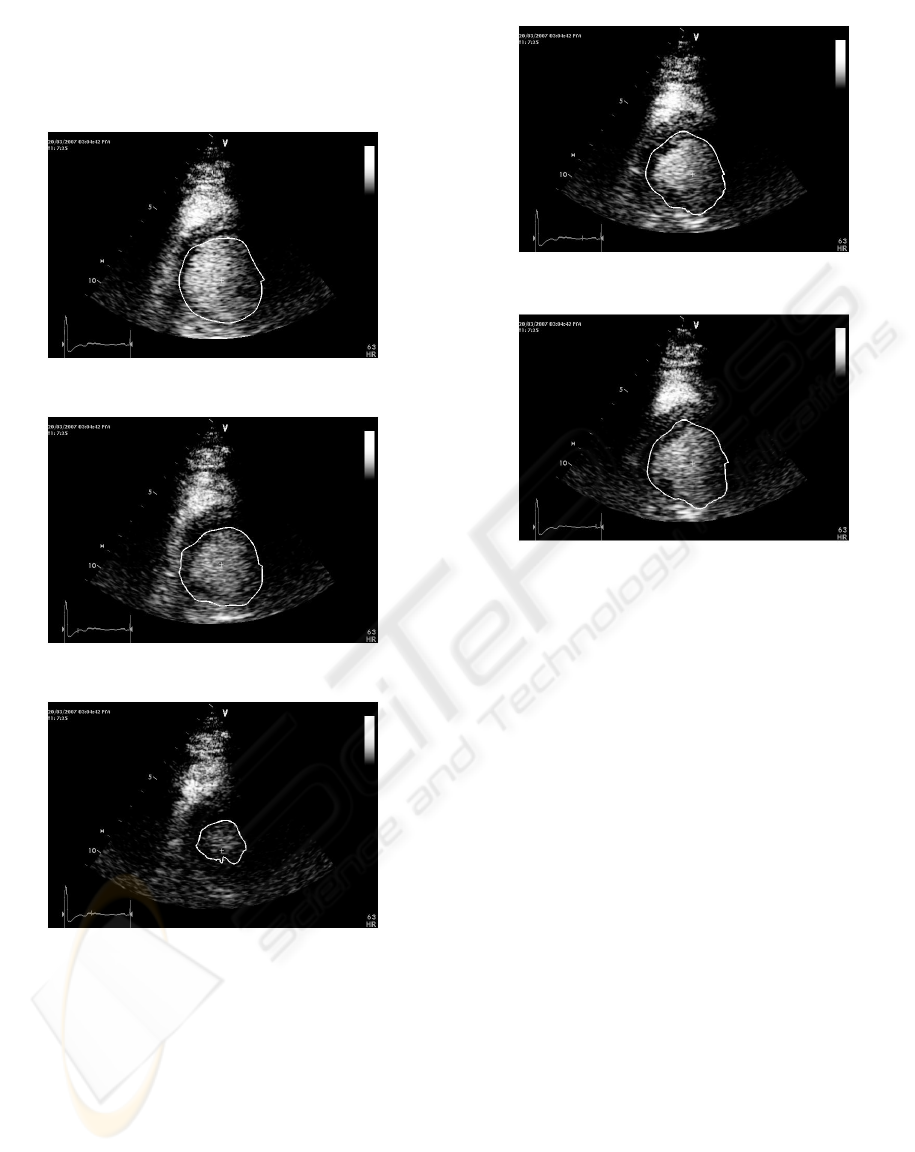
Figure 3: Detected contour in frame No.1.
Figure 4: Detected contour in frame No.8.
Figure 5: Detected contour in frame No.15.
4 EXPERIMENTAL RESULTS
The proposed methods for ellipse fitting, DBSCAN
and Spatio-temporal DBSCAN were implemented in
MATLAB 2006a on P-IV 2.1 GHz PC. Figures 3 to
7 show the result of application of the proposed algo-
rithm. The endocardial border estimation was done
on more than 10 video sequences of various standard
contrast echo views. The contour estimated by com-
puter in each frame of every sequence was compared
with that drawn by expert.
Figure 6: Detected contour in frame No.25.
Figure 7: Detected contour in frame No.32.
5 CONCLUSIONS
The proposed method for semi automatic estimation
of endocardial border of heart chambers in short axis
contrast echocardiographic sequences is based on el-
lipse fitting and subsequent spatio-temporal recursive
density-based clustering. The results show the effec-
tiveness of the method and its utility in the recovery
of the dropouts during image acquisition. The method
requires user intervention only in the first frame of
the sequence. The contour for each frame so obtained
may be utilized for the determination of the cardiac
parameters like, wall motion, area and for 3D visual-
ization. Further work is required before the method
can be employed in clinical environment for evalua-
tion of cardiac functional parameters. The issues in-
volved are the testing robustness, computational com-
plexity of the method along with its sensitivity to the
expert points. The algorithm requires fine tuning of
parameter λ for determination of optimum number of
iterations. In future work, we also intend to test the
proposed method on large number of data sets for its
further validation for images acquired in routine clin-
ical environment.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
208

ACKNOWLEDGEMENTS
The authors would like to thank Echocardiology de-
partment of Asian Heart Hospital, Mumbai for ex-
tending its support for contrast echo data and sugges-
tions during analysis and interpretation.
REFERENCES
S.L.Bridal, S.L. Correas, Saied, A. and Laugier, P.(2003)
Milestones on the road to higher resolution, quanti-
tative, and functional ultrasonic imaging. In Proc.
IEEE,vol.91, no.10, pp.1543-1561.
Maes, L. Bijnens, B. Suetens, P. and Verf, F.V. Automatic
Contour Detection of the Left Ventricle in short Axis
views in 2D Echocardiograms. In Machine vision and
Applications, Springer-Verlag., 6:1-9.
Noble, J.A. and Boukerroui, D.(2006). Ultrasound Image
Segmentation: A Survey. In IEEE Trans. Med. Imag.,
vol. 25, pp. 987-1010.
Setaredhan, S. K. and Soragham, J. J.,(1996) Automatic
Cardiac LV Boundary Detection and Tracking Using
Hybrid Fuzzy Temporal and Fuzzy Multiscale Edge
Detection. In IEEE Trans. Med. Imag., vol. 46, pp.
1364-1378.
JL Cohen et.al.(1998) Improved left ventricular endocardial
border delineation and opacification with optison, a
new echocardiographic contrast agent. In J Am Coll
Cardiol, 32, pp: 746:752.
Wolfer, J. Lee, SH. Sandelski, J. Summerscales, R. Soble, J.
and Roberge, J.(1999) Endocardial Border Detection
in Contrast Enhanced Echocardiographic Cineloops
using a Pulse Coupled Neural Networks. In IEEE
Conference on Computers in Cardiology, pp. 185-188.
Zwirn, G. Beeri, R. Gilon, D. and Akselrod, S.(2006).
Automatic Endocardial-Boundary Detection in Low
Mechanical-Index Contrast Echocardiography. In
IEEE Trans.on Biomedical Engg., Vol.53, No. 11,
pp:2310- 2322.
Mulet-Parada, M and Noble, J.A.(1998). 2D+T Acoustic
Boundary Detection in Echocardiography. In MIC-
CAI’98. LNCS 1496, pp. 806-813.
Choy, M.M. and Jin, J.S.(1998) Extracting endocardial Bor-
ders from sequential Echocardiographic Images. In
IEEE Engineering in Medicine and Biology, pp. 116-
121.
MA Morales, MA. Positano, V. Rodriguez, O. Passera, M.
Lombardi, M. and Rovai, D., (2002) Tracking of the
Left Ventricle in Contrast Enhanced Echocardiogra-
phy by Anisotropic Filtering and Active Contours Al-
gorithm. In IEEE Conference on Computers in Cardi-
ology, pp. 65-68.
Garcia,J. Rotger, D. Carreras, F. Leta, R. and Radeva, P.
(2003) Contrast Echography Segmentation and Track-
ing by Trained Deformable Models In IEEE Confer-
ence on Computers in Cardiology, pp. 173-176.
J.E. Pickard, J.E. R.L. Janiiczek, R.L. S.T. Acton, S.T. J.
Sklenar, J. J.A. Hossack, J.A. and S. Kaul, S.,(2004)
Segmentation of the Myocardium from Myocardial
Contrast Echocardiography. In IEEE Conference
on Signals, Systems and Computers, Volume 2, pp:
1616-1619.
Mishra, A. Dutta, P. Ghosh, M.K.(2003) A GA Based Ap-
proach for boundary Detection of Left Ventricle with
Echocardiographic Image Sequences, In Image and
vision Computing vol.21, pp. 967-976.
Celebi, M.E. Aslandogan, Y.A. and Bergstresser,
P.R.(2005) Mining biomedical Images with Density-
based Clustering. In Proceedings of IEEE Conference
on Information Technology: Coding and Comput-
ing(ITCC05).
Jain, A.K. Dubes, R.C. (1998). Algorithms for Clustering
Data. Prentice Hall, Upper Saddle River, New Jersy.
Fung, G. (2001) A Comprehensive Overview of Basic Clus-
tering Algorithms In Technical Report
Jain, A.K. Murty, M.N. Flynn, P.J.(1999). Data Clustering:
a Review. In ACM Surveys, 31(3):264-323.
Bradley, PS. Fayyad, U. and Reinna, C.(1998) Scaling
Clustering algorithms to large data bases.
Han,J. and Kamber, M. (1998) Data Mining: Concepts and
Techniques. Morgan Kaufmann, San Francisco, 2000.
Fedele F, Trambaiolo P, Magni G, De Castro S and Cacciotti
L, New modalities of regional and global left ventric-
ular function analysis;State of the art. In American
Journal of Cardiology, 81:49G-57G.
Yu, Y. and Acton, S.T., (2002) Speckle Reducing
Anisotropic Diffusion In IEEE Trans.Image Process-
ing.,vol 11,No.11, pp. 126-1270.
Fitzgibbon, A. Pilu, M. and Fisher, R.B. (1999) Direct Least
Square fitting of Ellipses, In Pattern Analysis and Ma-
chine Intelligence, vol.21, no.5.
ENDOCARDIAL SEGMENTATION IN CONTRAST ECHOCARDIOGRAPHY VIDEO WITH DENSITY BASED
SPATIO-TEMPORAL CLUSTERING
209
