
A DNA-INSPIRED ENCRYPTION METHODOLOGY FOR
SECURE, MOBILE AD-HOC NETWORKS (MANET)
Harry C. Shaw and Sayed Hussein
NASA Goddard Space Flight Center, Greenbelt, MD, USA
Department of Electrical and Computer Engineering, George Washington University, Washington, DC, USA
Keywords: Encryption, DNA computing, MANET, Biomimetic, Molecular Cryptography, Steganography,
Computational Biology.
Abstract: Molecular biology models such as DNA evolution can provide a basis for proprietary architectures that
achieve high degrees of diffusion and confusion and resistance to cryptanalysis. Proprietary encryption
products can serve both large and small applications and can exist at both application and network level.
This paper briefly outlines the basis of the proprietary encryption mechanism which uses the principles of
DNA replication and steganography (hidden word cryptography) to produce confidential data. The
foundation of the approach includes: organization of coded words and messages using base pairs organized
into genes, an expandable genome consisting of DNA-based chromosome keys, and a DNA-based message
encoding, replication, and evolution process. Such an encryption model provides “Security by Obscurity”.
1 INTRODUCTION
Mobile Ad-hoc Networks (MANET) require the
ability to distinguish trusted peers, and transmit and
receive information confidentially, yet tolerate the
ingress and egress of nodes on an unscheduled,
unpredictable basis. Because the networks by their
very nature are mobile, self-organizing and self
assembling, use of a Public Key Infrastructure
(PKI), X.509 certificates, RSA and nonce exchanges
becomes problematic if the ideal of MANET is to be
achieved. The use of evolutionary computing and a
DNA (Deoxyribonucleic acid) inspired approach are
key in developing true MANET architectures. Future
network organizations could include corporations,
retail outlets, financial institutions organized into
self-assembling MANETs of convenience, entering
and leaving the network as necessary. Such
networks might be better served by encryption
approaches not widely available to the public.
This paper presents a new encryption technique
which utilizes DNA-inspired coding, a dynamic
fitness algorithm and trust metric vision for ad-hoc
routing, and a rapidly evolving basis of encryption.
Because of the dynamic, evolutionary nature of this
approach, potential intruders must continually
intercept decoding instructions between source and
destination. Missing one generation of genome
decryption information seriously corrupts the
decryption process. Missing multiple generations
eventually renders previous decryption analyses
useless.
2 BACKGROUND OF DNA
CRYPTOGRAPHY
The use of DNA as a cryptographic medium is not
new. DNA encryption systems are one of the paths
taken in the field of molecular computing. Systems
using DNA as a one-time code pad (Gehani, 1999)
in a steganographic approach have been described.
An image compression –encryption system using a
DNA-based alphabet (Bourbakis, 1997) was
demonstrated including a genetic algorithm based
compression scheme. Schemes utilizing DNA
encryption utilizing dummy sequences of DNA have
been published (Leier, 2000). The steganographic
approach is highly desirable because DNA provides
a natural template for the hidden message approach
(Clelland, 1999). Clelland is a pioneer in this field. It
also appears in recent applications such as DNA
watermarks (Heider, 2007).
472
C. Shaw H. and Hussein S. (2008).
A DNA-INSPIRED ENCRYPTION METHODOLOGY FOR SECURE, MOBILE AD-HOC NETWORKS (MANET).
In Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing, pages 472-477
DOI: 10.5220/0001068204720477
Copyright
c
SciTePress

3 PROBLEM STATEMENT
Figure 1 displays a MANET routed message from
Jack to Jill routed at two different times, through
secure and potentially malicious nodes. A truly ad
hoc network permits routing in the presence of un-
trusted peers. In this case, message traffic is between
Jack and Jill. Nodes A, B and C are trustworthy
nodes at time t
1
and nodes α and β are potentially
malicious nodes. At time t
2
, the situation is reversed.
The problem of successful routing of messages
over potentially un-trusted nodes requires:
Routed messages arrive at the destination intact
Routed messages remain confidential in
transmission
Cryptanalysis of message traffic passing through
nodes other Jack or Jill is unlikely to be successful.
Nodes enter and leave the network at will.
Figure 1: MANET routed over secure nodes at t
1
(
___
) and
secure nodes at t
2
(---).
4 ENCRYPTION PROCESS
• Two or more users define a plaintext dictionary,
and a DNA based dictionary. The users define
the method by which plaintext is represented by
the four DNA bases. The DNA dictionary is the
is the source of messages and encryption keys
(chromosomes)
• Messages are pre-coded from plaintext into
DNA using a system of linear equations relating
word position in the message and the ordinal
position in the dictionary
• Chromosomes encrypt multiple permutations of
the message
• The permutations are tested for fitness and the
most fit permutation is selected for transmission
by the source.
• The recipient decrypts the message with the
same chromosomes
• The genome is expanded by mutating the
chromosomes with each other or with message
sequences.
The system is based upon operations upon words
and not individual characters. The only individual
characters that are encrypted are one character
words.
Users of the DNA encryption tool are endowed
with a starter genome which provides the equivalent
of a small dictionary for initiating messages, an
intended recipient capable of possessing a secret,
shared key, and a secret encryption/decryption
sequence to initiate communication. Chromosomes
are “long” compared to message sequences.
Let D represent a dictionary (lexicographically
ordered set) of all words such that D
0
represents the
first word in the dictionary and that sender and
receiver compose messages of W
i
words (genes). A
function U converts words to sequences of DNA
bases B
q
as shown below:
D
i -1
< D
i
< D
i + 1
∀ i < n
(1)
W
i
, ⊆ D
n
(2)
D
i
= U(W
i
,B
q
) (3)
There exists a one-to-one mapping between the
plaintext dictionary and DNA dictionary built from
B
q
={A,T,C,G} and. The binary coding for the bases
is shown in table 1. Note that A and T, and C and G
are inverses.
Table 1: DNA base coding.
Base Binary
value
Base Binary
value
Adenine 0011 Thymine 1100
Cytosine 1001 Guanine 0110
Given an alphabet of n characters, words of
character length m, each plaintext word codes into a
DNA word (gene) of x basepairs in length creating c
i
possible combinations of DNA words for each
plaintext word and Y total combinations DNA words
for the dictionary as shown below.
log
2
(n) = x (4)
c
i
= 2
(x*m)
(5)
Y = Σ c
i
, i=1,…,i
max
(6)
For n=8 with a character set consisting of
{a,e,i,o,u,n,s,t}, and m= 3, there would be 584 total
entries. Selected entries from such a dictionary are
shown in table 2. Sequences of nonsense words can
be inserted between plaintext words. As the
character set and character length increases, the
number of possible words (mostly nonsense words)
increases exponentially. Actual words can be padded
with interspersed nonsense words to increase
security. Figure 2 shows displays the maximum size
of the DNA dictionary for 8, 32 and 256 character
A DNA-INSPIRED ENCRYPTION METHODOLOGY FOR SECURE, MOBILE AD-HOC NETWORKS (MANET)
473
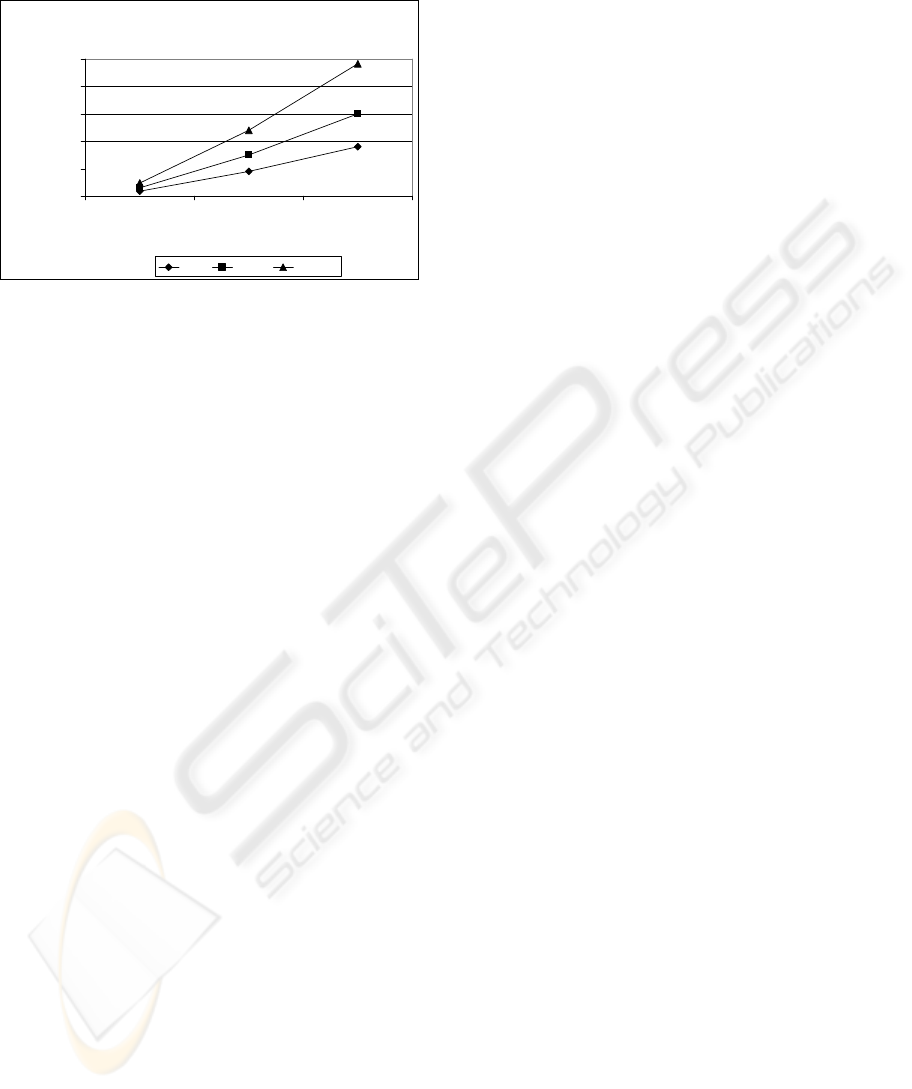
alphabets, word lengths ranging from 1 to 10
characters.
Maximum DNA Dictionary Size
1.E+00
1.E+05
1.E+10
1.E+15
1.E+20
1.E+25
1510
word length
words
Y(8) Y(32) Y(256)
Figure 2: DNA Dictionary Size.
Table 2: Sample DNA dictionary entries.
ordinal word DNA code
146 i TTT
147 ia TTACAT
148 ie TTATAT
452 san ACACACGCC
453 sas ACACAGAAG
454 sat ACACATAAT
455 sea ACCACACCC
The plaintext coding process yields message M
consisting of a sense string M
sense
of bases and M
anti-
sense
string of bases. Chromosome (C
1,..j
) sense and
anti-sense strands generated from the DNA
dictionary encrypt M
sense
and M
anti-sense
to produce
encrypted mutants. Given j chromosomes in the
genome, m message basepairs, k chromosome
basepairs, 2*j*(k-m) rounds of encryption on the
sense and anti-sense message strands are possible.
The message slides down the chromosome between
rounds. One encrypted mutant is produced per
round.
Table 3: Encryption Process.
Encrypt E(C,M) →Cipher
Anneal A(Cipher, B(q′)) → ACipher
Trust for p routes T
p
(FREQ
p
,RREQ
p
)→T
max
Fitness(Diffusion
& Confusion)
D(M,ACipher),C(M,ACipher)→ F
Select mutant S(g(F,T
max
)) → Output
Encryption is a 5 step process as shown in table
3. The encryption step processes the message against
the chromosome key to create a generation of two
new mutants: A DNA fragment consisting of a sense
strand from the message paired with a fragment of
equivalent length from the sense strand of each
chromosome moving from 5′to 3′ end, and a DNA
fragment consisting of an anti-sense strand from the
message paired with a fragment of equivalent length
from the sense strand of each chromosome key
moving from 3′to 5′ end. The process is summarized
in figure 4. The chromosome is depicted as a series
of segments. The functional output of the step is
referred to as ‘Cipher’
The process of aligning two dissimilar DNA
strands results in numerous mismatches. Figure 5
demonstrates the annealing process via
recombination and mutation by use of virtual bases
B′
q
= {a,t,c,g} Use of the transformation: A→g→T,
C→a→G, T→c→A, G→t→C simplifies the
evolution of the code and anneals mismatches.
Mutations are induced by the chromosome
(encryption key) onto the message (plaintext). The
rule is simple: if mismatch between the chromosome
base and a message base appears, the message is
mutated to match the chromosome. For example, if a
chromosome base ‘A’ is mismatched with either
‘A’, ‘C’ or ‘G’ on the corresponding message base,
the message base is changed to a ‘g’ which mutates
to a ‘T’. The unused bit patterns in the 4-bit binary
representations of the DNA hold special codes for
this transformation. The advantage of this technique
is that it allows for rapid merging of chromosome
and message strands and provides a path for
substituting new bases into the chromosome strand
and mimics the activity of creating molecules which
rely on the DNA structure but have substituted new
monomer units into the structure. This technique
could also be combined with crossover between
chromosome and message strand. The functional
output of this step is referred to as ‘ACipher’ for
annealed ciphertext.
The sender of the message would like to know
how much trust should be placed in each potential
route to the destination. Determining the level of
trust to be placed is a factor in determining the
fitness of the encryption. The source of trust
information in the methodology is querying the
network and tabulating successful forward and
return route request messages (FREQ, RREQ). The
value of this information decays between successive
queries. Given the assumption that any route is only
as secure as the weakest link, a trust metric for p
routes at a given point in time can be defined as:
T
p
(FREQ
p
,RREQ
p
,Δt)= (e-
Δt/r
)*ZF
p
*ZR
p
(7)
where ZF
p
and ZR
p
are the number of successful
forward and return route requests over p routes and
Δt is the delay from the baseline query. The rate of
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
474
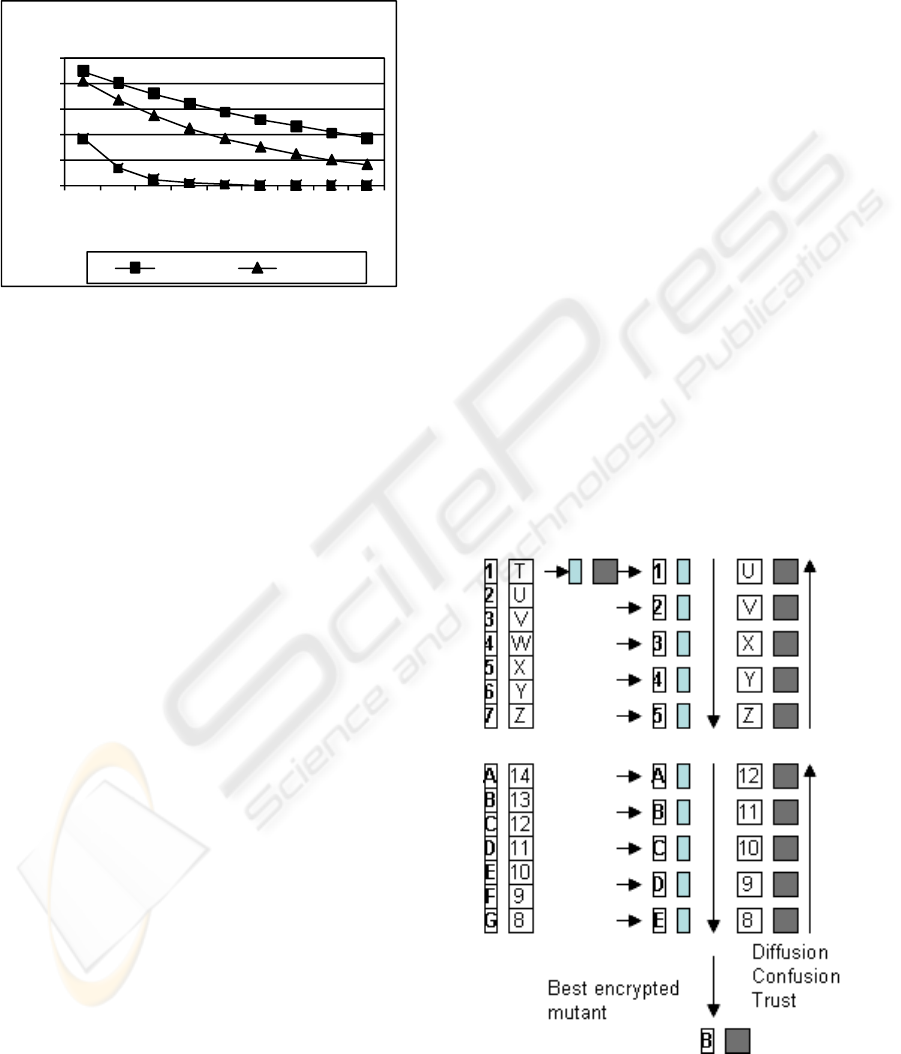
decay in trust can be adjusted by the factor r to
depending upon sender preference with the effects as
shown in figure 3. The maximum value from all T
p
represents the most desired route and is referred to
as ‘T
max
’.
0
0,2
0,4
0,6
0,8
1
0,1 0,2 0,3 0,4 0,5 0,6 0,7 0,8 0,9
trust
delta t
Dynamic Route Trust
r=0.9 r = 0.5
Figure 3: Temporal route trust.
A fitness algorithm defining the desired level of
diffusion and confusion (Shannon, 1949) produces
a means for evaluating each potential encryption.
Diffusion ensure that redundancy or patterns in the
plaintext message are dissipated into the long range
statistics of the ciphertext message. Confusion
ensures a complex relationship exists between the
plaintext and ciphertext. Each encrypted mutant is
compared to the plaintext message on this basis. The
output of these functions produces a fitness value,
‘F’, for each mutant.
The source can define a fitness goal, g(F,T
max
)
such that only an encrypted mutant that exceeds the
goal is selected by function S to become the
transmitted message, referred to as the ’ Output’.
Conceivably, if no mutant exceeded the fitness
goal, the sender could select one of the following
options:
a. Reduce the magnitude of the fitness
parameters diffusion and confusion
b. Query the network again, re-compute
T
max
and determine if there is an
encryption fit for transmission.
c. Conduct a second round of encryption
by mating the most fit encrypted
mutants, and re-compute their fitness
parameters
d. Delay transmission of the encrypted
message until a suitable T
max
is
achieved.
Figure 6 displays the transition from plaintext to
a pair of DNA strands 54 base pairs long (M
sense
and
M
anti-sense
) to a pair of encrypted and annealed
mutants with a sense strand from message and
chromosome for one mutant, and a anti-sense strand
from message and chromosme for a second mutant.
The 8 letter dictionary {a,e,i,o,u,n,s,t} and a
chromosome designated as C4 having 1793 base
pairs are used in this example.
5 MUTATION EFFECTS AND
FITNESS
Life is intolerant of a high mutation rate in its
genetic code. Ribonucleic acid (RNA) viruses have
the highest mutation rate of any living species, 10
-3
to 10
-5
errors/nucleotide and replication cycle
(Elena, 2006). The human DNA mutation rate has
been approximated to be on the order of 10
-8
errors/nucleotide and generation (Nachman, 2000).
Injection of mutations into DNA encrypted
messages is central to the encryption process.
In evolutionary biology, fitness is a characteristic
that relates to the number of offspring produced
from a given genome. From a population genetics
point of a view the relative fitness of the mutant
depends upon the number of descendants per wild-
type descendant. In evolutionary computing, a
fitness algorithm determines whether candidate
solutions, in this case encrypted messages, are
sufficiently encrypted to be transmitted.
Figure 4: Mating of chromosome to message and
subsequent selection.
A DNA-INSPIRED ENCRYPTION METHODOLOGY FOR SECURE, MOBILE AD-HOC NETWORKS (MANET)
475
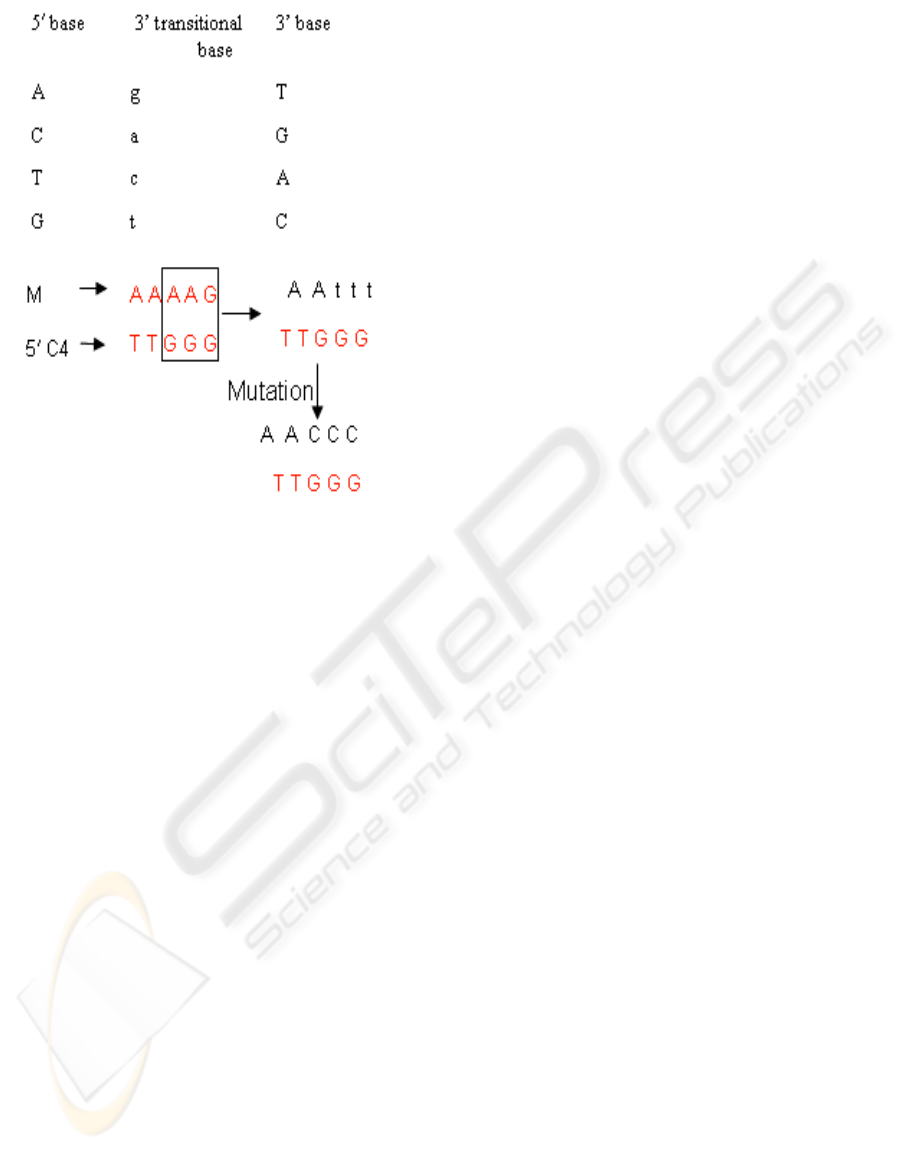
Figure 5: Anneal/mutation process.
By organizing the DNA dictionary into a codon
based system and applying tools of evolutionary
computing the encryption methodology can be
adapted as a tool of computational biology for
applications such as:
• Simulation of DNA mutations via
crossover and translation
• Creation of DNA samples and
mutagenic PCR (Polymerase Chain
Reaction) primers for simulation
• Optimization of alignment of two DNA
sequences
• Simulation of mutagenic agents on
DNA
• Rate-based synthesis and mutation
studies
Utilization of software based tools provides a
fast, cost-effective means of testing strategies prior
to performing laboratory analyses or cell-based
techniques. DNA coding for biological applications
require certain characteristics that are the opposite of
those required for encryption. Diffusion and
confusion must be minimized. Fitness would be
defined in application specific parameters such as
rate kinetics and reactant stoichiometry. Messages
could be replaced by oligonucleotides of interest.
6 CONCLUSIONS
A DNA-inspired encryption technique that is highly
resistant cryptographic analyses has been presented.
It is a new variation on an ancient cryptography
technique for use in mobile, ad-hoc networks and
does not require the use of a public key
infrastructure. To the best of the our knowledge, this
approach to providing confidentiality in a MANET
has not been previously published. The utility of a
rapid evolving encryption genome using transitional
annealing bases also represents a previously
unpublished concept.
Encryption users define the plain text dictionary,
the conversion into DNA sequences, the level of
trust to be conferred on the MANET and the fitness
characteristics of the message. The technique can be
used within MANETs without decryption to
establish cryptographic checksums for message
integrity, authentication, and secure electronic
transactions. It can be used within MANETs with
decryption for message confidentiality.
The methodology is extensible to the realm of
computational biology to perform computer aided
diagnostics of DNA mutations. It is also extensible
to other polymer based encryptions: peptide nucleic
acids, silicones, polysilanes, block co-polymers, etc.
It provides a path to simulating processes which
could be used for encoding messages into physical
molecules for a variety of applications.
ACKNOWLEDGEMENTS
This work was supported by NASA Goddard Space
Flight Center and the Space Communications and
Navigation Constellation Integration Project. Thanks
to Deborah M. Preston of DuPont Analytical
Solutions, Wilmington DE for reviewing this paper.
BIOSIGNALS 2008 - International Conference on Bio-inspired Systems and Signal Processing
476

Figure 6: Sample plaintext, encrypted mutation, annealed mutation.
REFERENCES
A Gehani, TH LaBean, JH Reif, 1999, DNA-Based
Cryptography, 5th DIMACS Workshop on DNA Based
Computers
Bourbakis, N.G., 1997, Image Data Compression-
Encryption Using G-Scan Patterns, Computational
Cybernetics and Simulation., 1997 IEEE International
Conference on
A Leier, C Richter, W Banzhaf, H Rauhe, 2000,
Cryptography with DNA binary strands, BioSystems
57, Elsevier
Catherine Taylor Clelland, Viviana Risca, Carter Bancroft,
1999, Hiding Messages in DNA microdots, Nature,
Macmillan
Dominik Heider and Angelika Barnekow, 2007, DNA-
based watermarks using the DNA-Crypt algorithm,
BMC Bioinformatics 2007, BioMed Central
Shannon, Claude, 1949, Communication Theory of
Secrecy Systems, Bell System Technical Journal
Santiago F Elena, Purificación Carrasco, José-Antonio
Daròs, Rafael Sanjuán. 2006, Mechanisms of genetic
robustness in RNA viruses. EMBO Report
Nachman MW, Crowell SL., 2000, Estimate of the
mutation rate per nucleotide in humans. Genetics,
Genetics Society of America
A DNA-INSPIRED ENCRYPTION METHODOLOGY FOR SECURE, MOBILE AD-HOC NETWORKS (MANET)
477
