
A LOSSLESS COMPRESSION ALGORITHM FOR DNA
SEQUENCES
Taysir H. A. Soliman
Faculty of Computer and Information, Assiut University, Egypt
Tarek F. Gharib, Alshaimaa Abo-Alian, Mohammed Alsharkawy
Faculty of Computer and Information Sciences, Ain Shams University, Egypt
Keywords: Lossless Compression Algorithm, encoding, approximate repeats, palindrome.
Abstract: Homology search is the seed for both genomics and proteomics research. However, the increase of the
amount of DNA sequences requires efficient computational algorithms for performing sequence comparison
and analysis. This is due to the fact that standard compression algorithms are not able to compress DNA
sequences because they do not consider special characteristics of DNA sequences (i.e. DNA sequences
contain several approximate repeats and complimentary palindromes are frequent in DNA). Recently, new
algorithms have been proposed to compress DNA sequences, often using detection of long approximate
repeats. The current work proposes a Lossless Compression Algorithm (LCA), providing a new encoding
method. LCA achieves a better compression ratio than that of existing DNA-oriented compression
algorithms, when compared to GenCompress and DNACompress, using nine different datasets.
1 INTRODUCTION
Molecular sequence databases (e.g., EMBL
(http://www.ebi.ac.uk/embl/), GenBank
(http://www.ncbi.nlm.nih.gov/Genbank/), Enterz
(http://www.ncbi.nlm.nih.gov/Entrez/), etc.)
currently collect terabytes of sequences of
nucleotides and amino-acids from biological
laboratories all over the world and are under
continuous expansion (Apostolico and Lonardi,
2000). These sequences play a vital role in
performing homology search to find motifs of
particular importance and to predict biomarkers.
DNA sequences contain only four bases {A, C, T,
G}. Thus, each base can be represented by two bits.
Some characteristics of DNA sequences show
that they are not random sequences (Behzadi and
Fessant, 2004). Two characteristic structures of
DNA sequences are known as approximate repeat
and complementary palindrome (Matsumuto,
Sadakane and Imai, 2000). Approximate repeat are
repeat with mutations i.e. a change in the DNA bases
"letters". Mutations are represented by edit
operations; Insert (I), Delete (D), Replace (R) and
the match or Copy represented by C (Tubingen and
Huson, 2005). Figure 1 shows the edit operations,
given two sequences: s
1
: “gaccgtcatt” and s
2
:
“gaccttcatt”.
(a) C C C C R C C C C C
g a c c g t c a t t
g a c c t t c a t t
Or C C C C D C I C C C C
(b) g a c c g t c a t t
g a c c t t c a t t
Figure 1: Examples of Séquence Mutations by Edit
Operations.
Complementary palindrome is a reversed repeat,
where A and C are respectively replaced by T and G
(Chen, Kwong and Li, 1999; Allison, Edgoose and
Dix, 1998). For example, consider the sequence
ACGCCT; its palindrome will be TCCGCA and its
complementary palindrome will be AGGCGT. The
effective handling of these features is the key to
successful DNA compression.
DNA sequences compression has recently
become a challenging computational problem.
Main advantages of compressing DNA sequences
435
H. A. Soliman T., F. Gharib T., Abo-Alian A. and Alsharkawy M. (2008).
A LOSSLESS COMPRESSION ALGORITHM FOR DNA SEQUENCES.
In Proceedings of the Tenth International Conference on Enterprise Information Systems - AIDSS, pages 435-441
DOI: 10.5220/0001683504350441
Copyright
c
SciTePress

arise from the available large and rapidly increasing
amount of DNA sequences. Moreover, data
compression now has an even more important role in
reducing the costs of data transmission, since the
DNA files are typically shared and distributed over
the Internet in heterogeneous databases. Space-
efficient representation of the data reduces the load
on FTP service providers such as GenBank.
Consequently, file transmissions are done faster,
saving costs for clients who access those files
(Korodi and Tabus, 2005). Furthermore, modeling
and analyzing DNA sequences may lead into
significant results, leading into finding a good
relatedness measurement between sequences may
lead into effective alignment and phylogenetic tree
construction. In addition, the statistical significance
of DNA sequences show how sensitive the genome
is to random changes, such as crossover and
mutation, what the average composition of a
sequence is, and where the important composition
changes occur (Korodi and Tabus, 2005).
Additionally, DNA compression has been used to
distinguish between coding and non-coding regions
of a DNA sequence, to evaluate the “distance”
between DNA sequences and to quantify how much
two organisms are “close” in the evolutionary tree,
and in other biological applications. On the other
hand, the standard text compression tools, such as
compress, gzip and bzip2, cannot compress these
DNA sequences since the size of the files encoded
with these tools is larger than two bits per symbol.
Data compression is a process that reduces the
data size. One of the most crucial issues of
classification is the possibility that the compression
algorithm removes some parts of data which cannot
be recovered during the decompression. The
algorithms removing permanently some parts of data
are called lossy, while others are called lossless
(Deorowicz, 2003).
In this paper, we propose a Lossless
Compression Algorithm (LCA) which consists of
three phases will be discussed in detail in section 4.
We use PatternHunter as a part of the first phase to
find approximate repeats and complementary
palindromes. LCA proposes a new encoding
methodology for DNA Compression. The rest of this
paper is organized as follows: in section 2, we
survey different algorithms for DNA sequence
compression. In section 3, the difference between
BLAST and PatternHunter is explained, illustrating
why PatternHunter is used to detect approximate
repeats and complementary palindromes in the
proposed algorithm. In section 4, our proposed
algorithm is explained. Section 5 shows the
comparison of our results on a standard set of DNA
sequences with results published for the most recent
DNA compression algorithms. Finally, section 6
presents the conclusion and future work.
2 RELATED WORK
Several compression algorithms have been
developed, such as Biocompress (Grumbach and
Tahi, 1993), Biocompress-2 (Grumbach and Tahi,
1994), C-fact (Rivals, Delahaye, Dauchet and
Delgrange, 1996), GenCompress (Chen, Kwong and
Li, 1999; Chen, Kwong and Li, 2001), CTW+LZ
(Matsumuto, Sadakane, and Imai, 2000),
DNASequitur (Cherniavski and Lander, 2004),
DNAPack (Behzadi and Fessant,, 2004), and
LUT+LZ (Bao, Chen and Jing, 2005). The first two
developed algorithm for compressing DNA
sequences are Biocompress, and its second version
Biocompress-2. They are similar to the Lempel-Ziv
data compression method in the way they search the
previously processed part of the sequence for
repeats. Biocompress-2 performs compression by
the following steps: 1) Detecting exact repeats and
complementary palindromes located in the already
encoded sequence and 2) Encoding them by the
repeat length and the position of a previous repeat
occurrence. In case of no significant repetition is
found, Biocompress-2 utilizes order-2 arithmetic
coding. Another algorithm is the Cfact algorithm,
which searches for the longest exact matching repeat
using a suffix tree on the entire sequence. By
performing two passes, repetitions are encoded when
the gain is guaranteed; otherwise the two-bits-per
base (2-Bits) encoding is used. The GenCompress
algorithm is a one-pass algorithm based on
approximate matching, where two variants exist:
GenCompress-1 and GenCompress-2.
GenCompress-1 uses the Hamming distance (only
Replace) for the repeats while GenCompress-2 uses
the edition distance (deletion, insertion and Replace)
for the encoding of the repeats. CTW+LZ
algorithm is based on the context tree weighting
method. It combines a LZ-77 type method like
GenCompress and the CTW algorithm. Long
exact/approximate repeats are encoded by LZ77-
type algorithm, while short repeats are encoded by
CTW. Although good compression ratios are
obtained, execution time is too high to be used for
long sequences. DNASequitur is a grammar-based
compression algorithm for DNA sequences which
infers a context-free grammar to represent the input
data but fails to achieve better compression than
ICEIS 2008 - International Conference on Enterprise Information Systems
436

other DNA Compressors. DNAPack uses Hamming
distance for computing the approximate repeats and
complementary palindromes, and either CTW or
order-2 arithmetic coding compression for the non-
repeat regions. Unlike the above algorithms,
DNAPack does not choose the repeats by a greedy
algorithm, but uses a dynamic programming
approach instead. LUT+LZ algorithm combines a
Lookup Table (LUT)-based pre-coding routine and
LZ77 compression routine. LUT maps the
combination of ATGC into 64 ASCII characters.
In addition, several DNA compression tools
exist, such as DNACompress (Chen, Li, Ma and
Tromp, 2002) and DNAC (Chang , 2004).
DNACompress is a DNA compression tool, which
employs the Lempel-Ziv compression scheme as
Biocompress-2 and GenCompress. It consists of two
phases: 1) Finding all approximate repeats including
complementary palindromes, using specific
software, PatternHunter, and 2) Encoding the
approximate repeats and the non-repeat regions. In
practice, the execution time of DNACompress is
much less than GenCompress. DNAC is another
DNA compression tool, working in four phases: 1)
Building a suffix tree to locate exact repeats, 2)
Extending approximate repeats, 3) Extracting the
optimal non-overlapping repeats from the
overlapping ones, and 4) Encoding all the repeats.
In the next section, a brief comparison between
PatternHunter and BLAST is performed.
3 PATTERNHUNTER V.S. BLAST
PatternHunter is a tool, which performs homology
search algorithm, using a novel seed model for
increased sensitivity and new hit-processing
techniques for significantly increased speed (Ma,
Tromp and Li, 2002).
PatternHunter finds all “approximate repeats”
and “complementary palindromes” in one DNA
sequence or between a pair of DNA sequences and
outputs all repeats ranked by score. Blast looks for
matches of k consecutive letters as seeds. Instead
PatternHunter uses non-consecutive k letters as
seeds. When comparing PatternHunter with BLAST
in terms of speed and memory usage, PatternHunter
performs better than BLAST, where PatternHunter
runs up to 20 times faster than BLAST. Yet, at the
same time, PatternHunter is more sensitive i.e. fewer
high-quality matches are missed by PatternHunter
than by BLAST (Ma, Tromp and Li, 2002).
For example, PatternHunter can search for
homologies between Arabidopsis Chromosome 2
(20 Mb) and Chromosome 4 (18 Mb) in under 15
minutes on a 700 MHz Pentium III with 1 GB of
main memory. BLAST cannot complete this task on
the same computer due to its high memory
requirements. PatternHunter was used by the Mouse
Genome Sequencing Consortium to compare human
and mouse genomes. The task was completed in 20
CPU-days; it was estimated that the same task would
take 20 CPU-years with BLAST
(http://www.bioinformaticssolutions.com/products/p
h/index.php).
4 LCA PHASES AND ANALYSIS
The proposed Lossless Compression Algorithm
(LCA) considers the special characteristics of DNA
sequences so it uses a new encoding method. LCA
consists of three main phases, as shown in Figure 2.
1) Finding approximate repeats and complementary
palindrome, 2) Removing overlapping between
repeats by choosing ones with high scores and
encoding repeats and palindromes after removing
overlapping, and 3) Encoding the non-repeat
regions.
4.1 Finding Approximate Repeats and
Complementary Palindromes
Because of sensitivity and efficiency of
PatternHunter, we use it to detect approximate
repeats and complementary palindromes in the input
DNA sequence in the first phase. Our algorithm uses
the standard parameters for PatternHunter. These
parameters were set and optimized based on the
number of bits needed to encode a mismatching base
as well as to encode a match. The output file of
PatternHunter contains information about each
repeat such as its score, start and end position, a set
of edit operations, and whether it is an approximate
repeat or a palindrome.
Figure 3 shows a sample of an output file of
PatternHunter using HUMVIR sequence as an input.
In the output file, the 'Sbjct' is the repeat segment
which will be encoded with respect to 'Query'.
‘Query’ and ‘sbjct’ are followed by their start
positions, sequence of bases, and end positions.
A LOSSLESS COMPRESSION ALGORITHM FOR DNA SEQUENCES
437
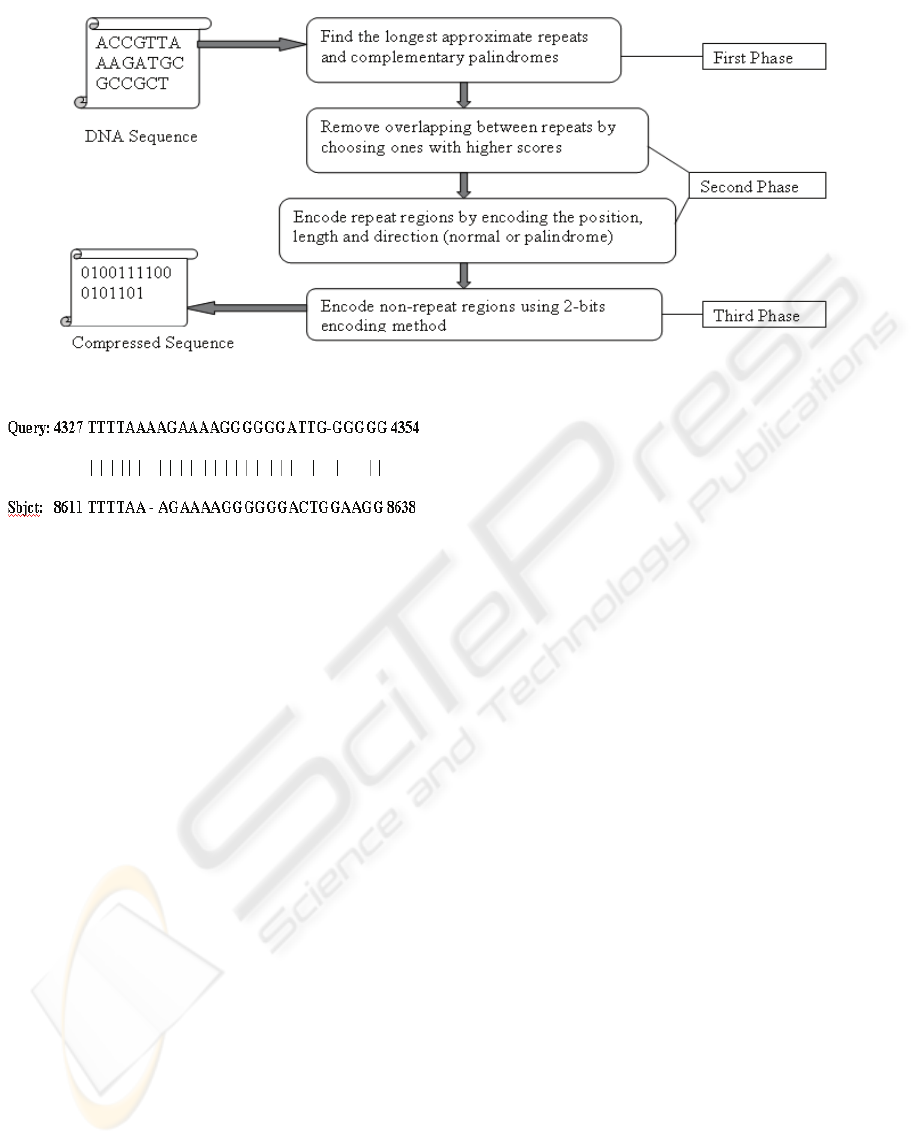
Figure 2: Phases of proposed DNA compression Algorithm.
Figure 3: A sample of an output file of PatternHunter.
The '|' means there is a Copy or Match operation.
The '-' when exists in query, it means there is
insertion operation but when exists in sbjct; it means
there is deletion operation. When an end position of
a query is less than its start position, it means that its
subject is a complementary palindrome. Otherwise,
it is an approximate repeat.
4.2 Encoding Repeat Regions
The second phase develops the new encoding
scheme which is the main contribution of this paper.
In this phase, we first remove overlapping between
repeats by choosing one with higher score. Then, we
encode each repeat, using the following encoding
scheme:
The algorithm uses two bits to determine
whether the following block of bits is
approximate repeat, complementary
palindrome, edit operation, or non-repeat
segment.
o 01: an approximate repeat, followed
by start position of query and the
length.
o 10: a complementary palindrome,
followed by start position of
query and the length.
o 11: edit operations, followed by
number of edit operations, the
code of each operation.
o 00: non-repeat segment.
When encoding numbers in case of position or
length, the algorithm searches for the
maximum position to get the fixed number of
bits to represent each position. Then, the
algorithm converts the number to its binary
representation.
The algorithm uses two bits to encode each edit
operation:
o Replace (R): 00 followed by its
relative position and the code of
base to be replaced.
o Insertion (I): 01 followed by its
relative position and the code of
base to be inserted.
o Deletion (D): 10 followed by its
relative position.
4.3 Encoding Non-Repeat Regions
In the third phase, we extract non-repeat segments
from the DNA sequence and encode them using 2-
bit encoding method which replaces each base with
two bits as the following A (00), C (01), G (10), and
T (11). Figure 4 summarizes different steps of the
proposed algorithm.
ICEIS 2008 - International Conference on Enterprise Information Systems
438
5 EXPERIMENTAL RESULTS
AND DISCUSSION
We compare the results of our algorithm to the most
recent DNA compression algorithms. These
experiments are performed on a computer whose
CPU is Pentium IV 3GHz, memory is 512 MB and
OS is Windows XP SP2. Table 1 shows the
compression ratios (the number of bits per base) of
these algorithms on standard benchmark sequences.
These sequences are downloaded from NCBI
database in FASTA format. Table 2 shows a short
description about these sequences. Table 3 compares
running time of these algorithms. LCA seems to be
similar to DNACompress. However, LCA relies on
different encoding methods for repeat and non-
repeat regions. Our program achieves better
compression ratios than other programs except in
three cases, as shown in Table 1. Although we failed
to compress HUMDYSTROP efficiently because its
approximate repeats are infrequent, the execution
time of LCA is 1.33 seconds. HEHCMVCG and
VACCG contain approximate repeats with many
edit operations so we can not achieve the optimal
compression ratio for them.
The complexity of LCA is O( s (p + n) ) where s
is the number of approximate repeats and
complementary palindrome segments in the input
DNA sequence, p is the number of edit operations in
each segment, and n is the length of the input DNA
sequence.
6 CONCLUSIONS AND FUTURE
WORK
DNA compression computation is getting a lot of
attention. However, existing algorithms do not prove
good compression ratios because of the
characteristics of DNA sequences. In this work, we
proposed LCA for compressing DNA sequences,
relying on encoding methods, where its other phases
are similar to existing algorithms. LCA proved to
have better compression algorithms, when compared
to other two algorithms: GenCompress and
DNACompress. Nine different datasets have been
used: HUMGHCSA, HUMHPRTB,
HUMDYSTROP, HUMHDABCD, HEHCMVCG,
CHMPXX, CHNTXX, MPOMTCG, and VACCG.
LCA proves to have better compression ratios than
the other algorithms using all datasets, except for
HUMDYSTROP, HEHCMVCG and VACCG
sequences. In future work, we will use our
compression algorithm to predict coding and non-
coding regions in DNA sequences.
Figure 4: The pseudocode of the proposed algorithm.
REFERENCES
Allison, L., Edgoose, T., Dix, T. I. (1998) ‘Compression
of strings with approximate repeats’, In Intelligent
Systems in Mol. Biol., 8–16, Montreal.
1. Get a set of the longest
approximate repeats and
complementary palindromes Segments.
2. Remove overlapping between
segments by discarding whose score
is lower than its overlapped
segment.
3. Encoding segments:
For each s in Segments
Begin
IF s is palindrome THEN
Scode = “10”;
Else
Scode = “01”;
Scode = Scode +
BinaryCode(s.startpos) +
Binarycode(s.length);
For each op in s.editoperations
Begin
Scode = Scode + code(op) +
BinaryCode (op.pos);
IF op.type is not ‘delete’
THEN
Scode = Scode + code
(op.base);
End
End
4. Encoding Non-repeat regions :
For each block b is not in
Segments
Begin
Bcode = “”;
For each base in b
Begin
Switch (base)
Begin
Case “A”:
Bcode = Bcode + “00”;
Case “C”:
Bcode = Bcode + “01”;
Case “G”:
Bcode = Bcode + “10”;
Case “T”:
Bcode = Bcode + “11”;
End
End
End
A LOSSLESS COMPRESSION ALGORITHM FOR DNA SEQUENCES
439

Apostolico A. and Lonardi S. (2000) ‘Compression of
Biological Sequences by Greedy Offline Textual
Substitution’, In proc. Data Compression Conference,
IEEE Computer Society Press, 143-152.
Bao, S., Chen, S., and Jing, Z. (2005) ‘A DNA Sequence
Compression Algorithm Based on Look-up Table and
LZ77’, Signal Processing and Information
Technology, Proceedings of the Fifth IEEE
International Symposium, 23 - 28.
Behzadi, B. and Le Fessant, F. (2004) ‘DNA Compression
Challenge Revisited’, Lecture Notes in Computer
Science 3537, 190-200.
Chang C.-H. (2004) ‘DNAC: A Compression Algorithm
for DNA Sequences by Nonoverlapping Approximate
Repeats’, Master Thesis.
Cherniavski, N., Lander, R. (2004) ‘Grammar-based
Compression of DNA sequences’, in DIMACS
Working Group on The Burrows—Wheeler
Transform, Piscataway, NJ, USA.
Chen, X., Kwong, S., Li, M. (1999) ‘A compression
Algorithm for DNA sequences and its applications in
genome comparison’, The 10th workshop on Genome
Informatics, 51-61, Tokyo, Japan.
Chen, X., Kwong, S., Li, M. (2001) ‘A compression
Algorithm for DNA sequences’, IEEE Engineering in
Medicine and Biology Magazine, 20(4), 61-66.
Chen, X., Li, M., Ma, B. and Tromp, J. (2002)
‘DNACompress: fast and effective DNA sequence
compression’, Bioinformatics, 18, 1696-1698.
Deorowicz, S. (2003) ‘Universal lossless data compression
algorithms’, Philosophy Dissertation Thesis, Gliwice.
Grumbach S. and Tahi F. (1993) ‘Compression of DNA
Sequences’, In Data compression conference, IEEE
Computer Society Press, 340-350.
Grumbach S. and Tahi F. (1994) ‘A new Challenge for
compression algorithms: genetic sequences’, Journal
of Information Processing and Management, 30, 875-
866.
Korodi, G., Tabus, I. (2005) ‘An Efficient Normalized
Maximum Likelihood Algorithm for DNA Sequence
Compression’, ACM Transactions on Information
Systems, 23(1), 3–34.
Ma, B., Tromp, J., Li, M. (2002) ‘PatternHunter–faster
and more sensitive homology search’, Bioinformatics,
18, 440-445.
Matsumuto, T., Sadakane, K.,Imai, H. (2000) ‘Biological
sequence compression algorithms’, Genome Inform.
Ser. Workshop Genome Inform, 11, 43-52.
Rivals E., Delahaye J.-P., Dauchet M., Delgrange O.
(1996) ‘A Guaranteed Compression Scheme for
Repetitive DNA Sequences’, Data Compression
Conference, 453, Snowbird,
Tubingen, U., Huson, D. (2005) ‘Sequence comparison by
compression’, Alg. in Bioinformatics I, ZBIT, 18, 1-8.
http://www.bioinformaticssolutions.com/products/ph/inde
x.php
http://www.ebi.ac.uk/embl/
http://www.ncbi.nlm.nih.gov/Entrez/
http://www.ncbi.nlm.nih.gov/Genbank/
ICEIS 2008 - International Conference on Enterprise Information Systems
440
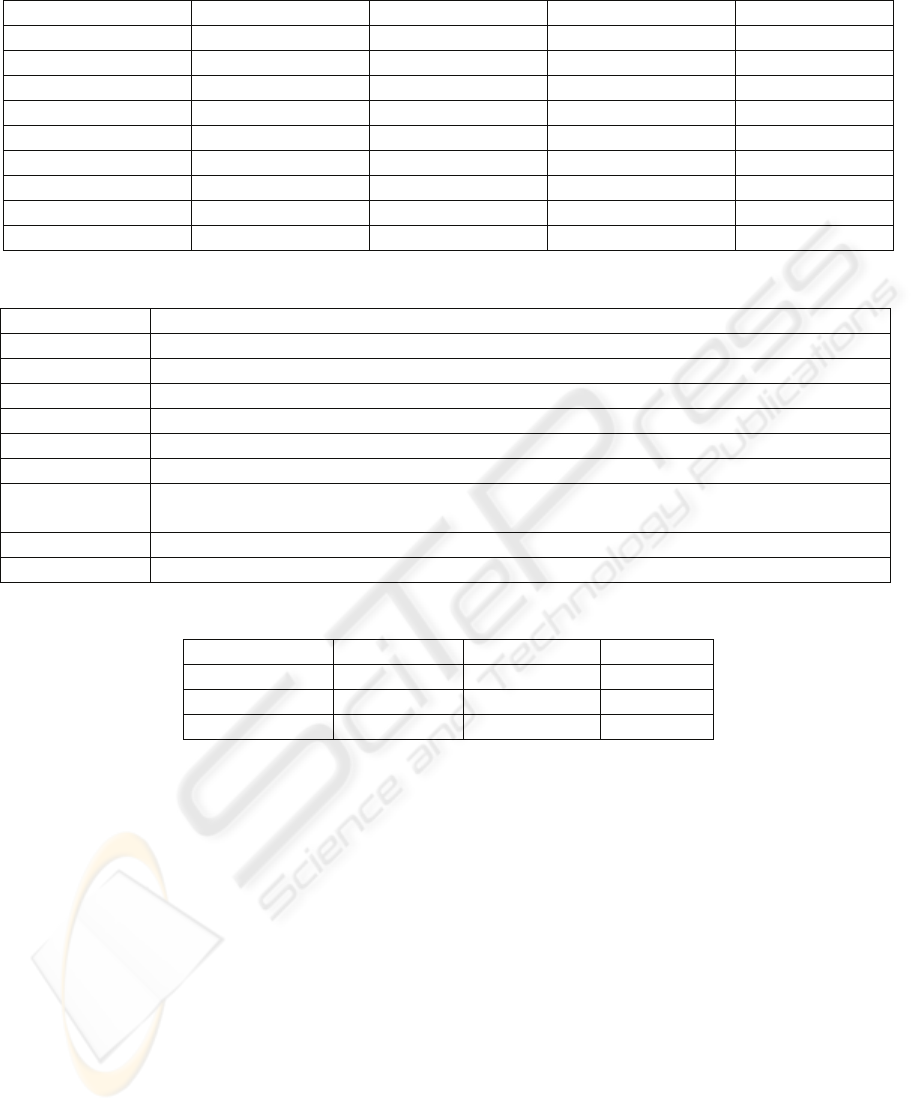
Table 1: Comparison of compression ratios for different algorithms (bits/base).
Table 2: Description about the datasets.
Table 3: Comparison of running times in seconds.
DNA Sequence GenCompress DNACompress LCA
HUMDYSTROP 6 2 1.33
HEHCMVCG 51 3.4 3
HUMHDABCD 11 2.5 2
Sequence Length (bases) GenCompress DNACompress LCA
HUMGHCSA 66495 1.0969 1.0272 1.0216
HUMHPRTB 56737 1.8466 1.8165 1.7911
HUMDYSTROP 38770 1.9231 1.9116 2.0113
HUMHDABCD 58864 1.8192 1.7951 1.7569
HEHCMVCG 229354 1.847 1.8492 1.9617
CHMPXX 121024 1.673 1.6716 1.613
CHNTXX 155844 1.6146 1.6127 1.6018
MPOMTCG 186609 1.9058 1.892 1.8849
VACCG 191737 1.7614 1.7580 1.7601
Description DNA Sequence
Human growth hormone and chorionic somatomammotropin genes HUMGHCSA
Human hypoxanthine phosphoribosyltransferase (HPRT) gene HUMHPRTB
Human dystrophin gene HUMDYSTROP
Human DNA sequence of contig comprising 3 cosmids (HDAB, HDAD, HDAC) HUMHDABCD
Human cytomegalovirus , a betaherpesvirus, represents the major infectious cause of birth defects HEHCMVCG
the complete chromosome III from yeast CHMPXX
Nectary tissues from flowers at Stage 6 of development from mature greenhouse grown Nicotiana
langsdorffii X Nicotiana sanderae (LxS8) plants
CHNTXX
Marchantia polymorpha mitochondrion MPOMTCG
Vaccinia virus Copenhagen VACCG
A LOSSLESS COMPRESSION ALGORITHM FOR DNA SEQUENCES
441
