
Ontology-driven Vaccination Information Extraction
Liliana Ferreira
1
, Ant
´
onio Teixeira
1,2
and Jo
˜
ao Paulo Silva Cunha
1,2
1
Institute of Electronics and Telematics Engineering of Aveiro
Campus Universit
´
ario de Santiago
3810-193 Aveiro, Portugal
2
Department of Electronics, Telecommunications and Informatics
Campus Universit
´
ario de Santiago
3810-193 Aveiro, Portugal
Abstract. Increasingly, medical institutions have access to clinical information
through computers. The need to process and manage the large amount of data
is motivating the recent interest in semantic approaches. Data regarding vacci-
nation records is a common in such systems. Also, being vaccination is a ma-
jor area of concern in health policies, numerous information is available in the
form of clinical guidelines. However, the information in these guidelines may
be difficult to access and apply to a specific patient during consultation. The cre-
ation of computer interpretable representations allows the development of clinical
decision support systems, improving patient care with the reduction of medical
errors, increased safety and satisfaction. This paper describes the method used
to model and populate a vaccination ontology and the system which recognizes
vaccination information on medical texts.The system identifies relevant entities
on medical texts and populates an ontology with new instances of classes. An
approach to automatically extract information regarding inter-class relationships
using association rule mining is suggested.
1 Introduction
Electronic access to clinical information in healthcare institutions is becoming a gen-
eralized reality through numerous Electronic Patient Record (EPR) systems. The need
to manage the increasingly large amount of patient data is motivating the recent in-
terest in semantic approaches, whose main goals are reducing medical errors, improve
physician efficiency and improve patient safety and satisfaction in medical practice. Se-
mantic Web technology helps achieve these goals using multiple populated ontologies,
automatic semantic annotation of documents, and rule processing [15].
Terms in medical domain are controlled leading to strict usage and less ambiguity.
Being this a strong advantage for a precise natural language processing, many work has
been developed in the area of medical language understanding. Medical language un-
derstanding aims at extracting the information content of medical texts. The information
representation may take the appearance of information formats filled with expressions
of the text [6] [12], or of a conceptual representation [15] [14]. In the set of studies con-
ducted for ontology development some obtained practical results like UMLS [17] and
SNOMED [16] but rely mainly on hand-made approaches. On the other hand, many
Ferreira L., Teixeira A. and Paulo Silva Cunha J. (2008).
Ontology-driven Vaccination Information Extraction.
In Proceedings of the 5th International Workshop on Natural Language Processing and Cognitive Science, pages 94-103
DOI: 10.5220/0001739500940103
Copyright
c
SciTePress

studies concentrate on full/semi-automatical techniques for knowledge-base building.
Serban et al. present in [13] a method to extract linguistic patterns with the purpose of
modeling breast cancer treatment guidelines, to aid the human modelers in guideline
formalization and reduce the human modeling effort.
This paper focuses on the introduction of semantic web technologies on an EPR
system towards a comprehensive system able to process patient data. The study con-
centrates on the conceptual representation of the portuguese vaccination guideline texts.
Vaccination is a major area of concern in health policies for which a lot of information
is available in the form of clinical guidelines. However, the information in these guide-
lines may be difficult to access and apply to a specific patient during consultation. The
creation of computer interpretable representations allows the development of clinical
decision support systems, improving patient care with the reduction of medical errors,
increased safety and satisfaction. The publicly availability of vaccination guidelines in
portuguese and the existence of a EPR system containing information about vaccination
records were the main motivation. An approach for creating a vaccine adverse event on-
tology has been reported by [7]. The concept modeling was carried out using Universal
Modeling Language (UML), from structured case definitions and the UMLS concept
table. However, as we intent to use the ontology in an EPR system in portuguese there
is need to model the Portuguese vaccination plan and so define a new knowledge base,
which, to our knowledge, has not been yet performed.
The work described in this paper concerns the development of populated ontologies
in the healthcare domain, specifically in the vaccination area and the development of
decision support algorithms that support rule and ontology based checking/validation
and evaluation.
The remainder of this paper is organized as follows. Section 2 introduces the EPR
system used in this study. The method used to model the vaccination guideline is pre-
sented on Section 3 and Section 4 describes the system developed to extract informa-
tions and populate the vaccination ontology. Section 5 highlights the main results and
innovation achieved with this approach and Section 6 concludes with futures work.
2 Motivating Scenario: RTS Project
In the last years a significant effort was invested in systems for electronic access to
clinical information within hospitals. This kind of access to clinical information is be-
coming a generalized reality in Europe through numerous enterprise-wide Electronic
Patient Record (EPR) systems. However, little effort was invested in systems for clini-
cal electronic communication between different medical institutions [2].
The ”Rede Telem
´
atica da Sa
´
ude” (RTS) project
3
is developing a telematic infras-
tructure to progressively support multiple clinical communication services at a regional
level. The project aims at promoting the secure electronic access to clinical informa-
tion stored in the different health care providers to all credentialed professionals. The
RTS provides a summarized Regional Electronic Clinic File that combines the clini-
cal electronic files existing in all the institutions that compose the network, smoothing
3
www.rtsaude.org
95

the clinical communication between different health institutions. The RTS allows in-
formation, such as release forms, clinical tests bulletins and vaccination records to be
accessible to the professionals from other institutions.
At the same time, citizens can manage their health issues over the RTS telematic
platform. Registered users can interact with their family doctor, request appointments
and manage their health agenda through the restricted area MyRTS. A public area is
also available with general interest health related contents, such as health guidelines
and health issues.
At the present time, the RTS provides access to more than 11.000.000 clinical
episodes from more than 35.000 patients. Several benefits have been experienced until
now, like avoiding duplication of diagnosis auxiliary exams or reducing unnecessary
patients’ visits.
However, most of the information available is in textual form and, even if elec-
tronically available, remains locked up within text. Enriching these systems with rich
domain knowledge and rules would greatly enhance their performance and ability to
support clinical decisions. The most important benefit we seek with the use of domain
knowledge like ontologies and rules is the reduction of medical errors through the de-
velopment of a clinical decision support system and better patient care with increased
safety and satisfaction.
3 Knowledge and Rules Representation
To develop the vaccination ontology we used the Portuguese Vaccination Program
(PVP) [5] as guideline. This program is publicly available and contains an elaborate
and systematic vaccination plan with information about chronological schemes, with
minimal and maximal age for the administration of each vaccine, information about the
dosages recommended, the anatomic places and the way of administration. Information
about possible reactions and several physical and drug interactions is also included.
To develop a formal description of the area we analyzed the guideline and manually
identified some linguistic patterns based on regularities in the text, such as medical
specific categories (disease, body
part, interaction) and lexical terms corresponding to
semantic relationships between medical categories. For instance, in the sentence
(1) At birth, it is recommended the vaccine against tuberculosis (BCG) and the first
dosage of the vaccine against hepatitis B (VHB), provided that the weight of the
new-born is greater than or equal to 2000 g.
4
the pattern
(2) AT [age] [vaccine] {against} [disease] ([acronym]) AND [dosage] [vaccine]
{against} [disease] ([acronym]), {restriction} [weight].
was identified.
Following this method we developed a formal description which models all the
concepts and relationships between concepts related with the vaccination area. This
description was used to create an ontology containing all of the vaccines and classes of
vaccines, vaccines interactions, and vaccines allergies.
4
Translation made by the author.
96
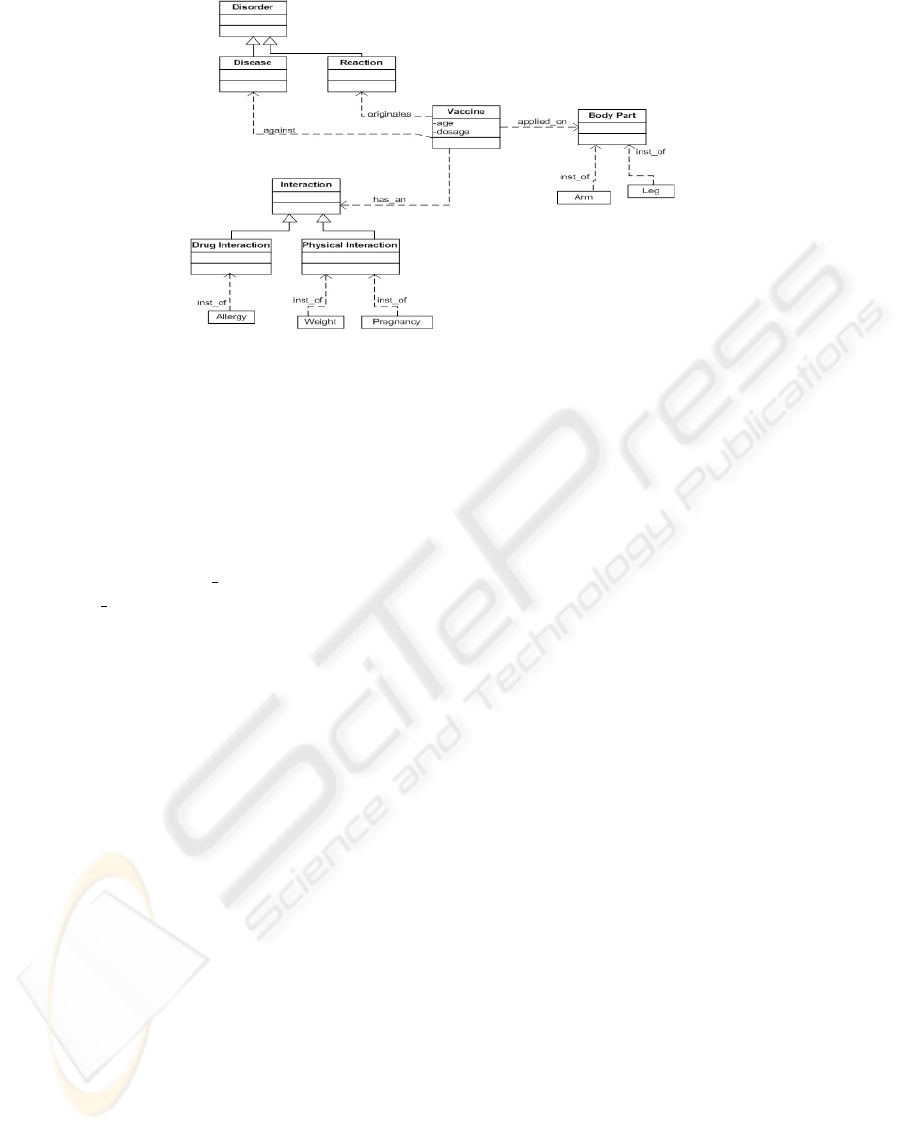
Fig. 1. Partial view of Vaccination Ontology.
A partial view of the ontology can be observed in Figure 1, which includes the
class Vaccine and it relationships with the classes Disease and Reaction, subclasses of
Disorder, with the class Interaction (subclasses Physical and Drug Interaction) and with
the class BodyPart.
In order to enrich the documents with domain knowledge we developed an OWL [11]
ontology. For each relationship between two class the inverse relation was also defined.
For instance, for the classes Vaccine and Disease was defined not only the relation-
ship Vaccine is against Disease but also the corresponding inverse relation Disease
is prevented Vaccine The ontology population was divided in two phases described in
Sections 4.1 and 4.2. The last step was to combine the OWL populated ontology with
SWRL [8] rules. Combining the use of ontology and rules, not only allows the system
to make decisions, but we can also extent the ontology with additional relationships and
facts without changing the code. Example rules include prevention of vaccine interac-
tion, for instance with a component of the vaccine (e.g., allergy interaction check) or
with a physical condition of the patient (e.g., pregnancy).Even though the rule specifi-
cation is at the moment very simple and has a preliminary character, it already provides
additional knowledge, for instance, allowing for better and more efficient patient care
introducing the possibility of offering suggestions when rules are broken or exceptions
made. Discussion of SWRL rules employment is not performed in the context of this
paper.
4 System Overview
The system developed to carry out the task of automatically extracting information from
the vaccination guidelines is derived from GATE [4] and has already been used with
the purpose of providing a formal description of Portuguese neurological reports [6].
GATE is an infrastructure for developing and deploying software components that pro-
cess human language, in development at the University of Sheffield since 1995. The
architecture consists of a pipeline of processing resources which run in series. Many
of these processing resources are domain-independent and can be used for Portuguese
97
(e.g., Tokenizer and Sentence Splitter). However, POS tagging and the main process-
ing, carried out by a gazetteer and by a set of grammar rules, had to be changed and
enriched with language and domain-specific parameters. A more detailed description is
given in [6].
In this study an improvement was made in order to automatically extract the in-
formation existing in the PVP. Ontology population was divided in two phases. First,
information regarding concepts modeled in the ontology was automatically extracted
and added in the ontology as instances of the corresponding classes. Finally, the inter-
instance relationships were manually added as OWL properties. An experiment with the
purpose of automatically performing this last step using an approach based on frequent
pattern mining was also performed. Section 4.1 describes the process of automatically
extracting information from PVP and populating the ontology, while Section 4.2 fo-
cuses on adding knowledge to the ontology.
4.1 Information Extraction
In order to automatically extract information related with the concepts defined on the
ontology a set of grammar rules were define. GATEs IE system is rule-based and re-
quires a developer to manually create rules, so it is not totally dynamic. The grammar
rules developed are written in JAPE (Java Annotations Pattern Language) [3]. The rules
do not just match instances from the gazetteer with their occurrences in the text, but
also find new instances in the text which do not exist in the gazetteer, through use of
contextual patterns, part-of-speech tags and other indicators.
Table 1. NER Annotation Set.
Category Type
VACCINE VACCINE
DISEASE DISORDER
REACTION DISORDER
ALLERGY DRUG INTERACTION
BODYPART BODYPART
AGE VACCINE
DOSAGE VACCINE
WEIGHT PHYSICAL INTERACTION
The entities to be identified for this task are in Table 1 and include the concepts of
Vaccine, Disease and Reaction identified as belonging to type Disorder their superclass,
Allergy and Weight as examples of Drug and Physical Interactions, respectively, and
Body Part. Information regarding Age and Dosage, properties of the class Vaccine, was
also extracted.
Figure 2 presents Gate’s Graphical Interface with an excerpt of the PVP automat-
ically annotated with the entities described above. The evaluation of this task is per-
formed in Section 5.
98
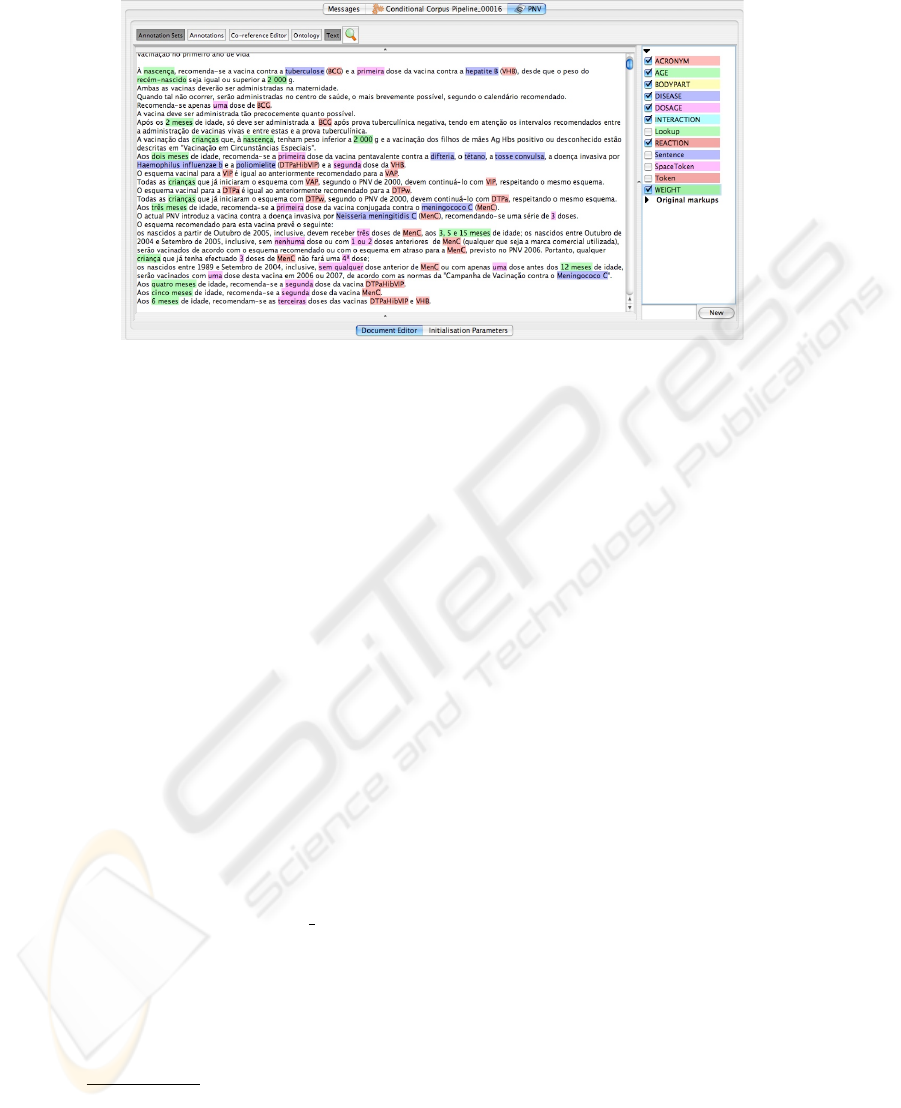
Fig. 2. Entities annotated on the Portuguese Vaccination Plan.
4.2 Populating the Ontology
The task of ontology population was divided in two steps. In the first step the informa-
tion about the entities extracted was automatically added in the ontology. In the second
step, the inter-instances relationships were filled. An attempt to automatize this step is
described.
Individuals. In the current scenario we have already an ontology and information ex-
tracted automatically and we want to populate it with instances whenever entities be-
longing to classes in the ontology are mentioned in the input texts. For that purpose a set
of grammar rules were developed. These match each annotation of indicated types and
the annotation span is used to extract the text covered in the document. Once all these
pieces of information are available, the addition to the ontology is performed. First the
right class in the ontology is identified using the class name and then a new instance for
that class is created. Figure 3 presents class Disease of the ontology populated with the
instances extracted from the text.
Relationships between Individuals. On a first approach inter-instance relationships
were manually added as OWL properties, using Prot
´
eg
´
e 3.4
5
. For instance, for the
acronym BCG, automatically populated on the ontology as an individual of the class
Vaccine, the relationship is against Tuberculose (portuguese word for Tuberculosis),
individual of class Disease, was defined. The same operation was done for the rest of
the individuals.
A first attempt towards an automatization of this step, using an approach based on
frequent pattern mining, was performed. For that we mined the annotated text for fre-
quent entity patterns, using Association Rule Mining [1]. Association rules describe
how often entities are mentioned together. For example, if the rule ’disease ⇒ vaccine
5
http://protege.stanford.edu
99
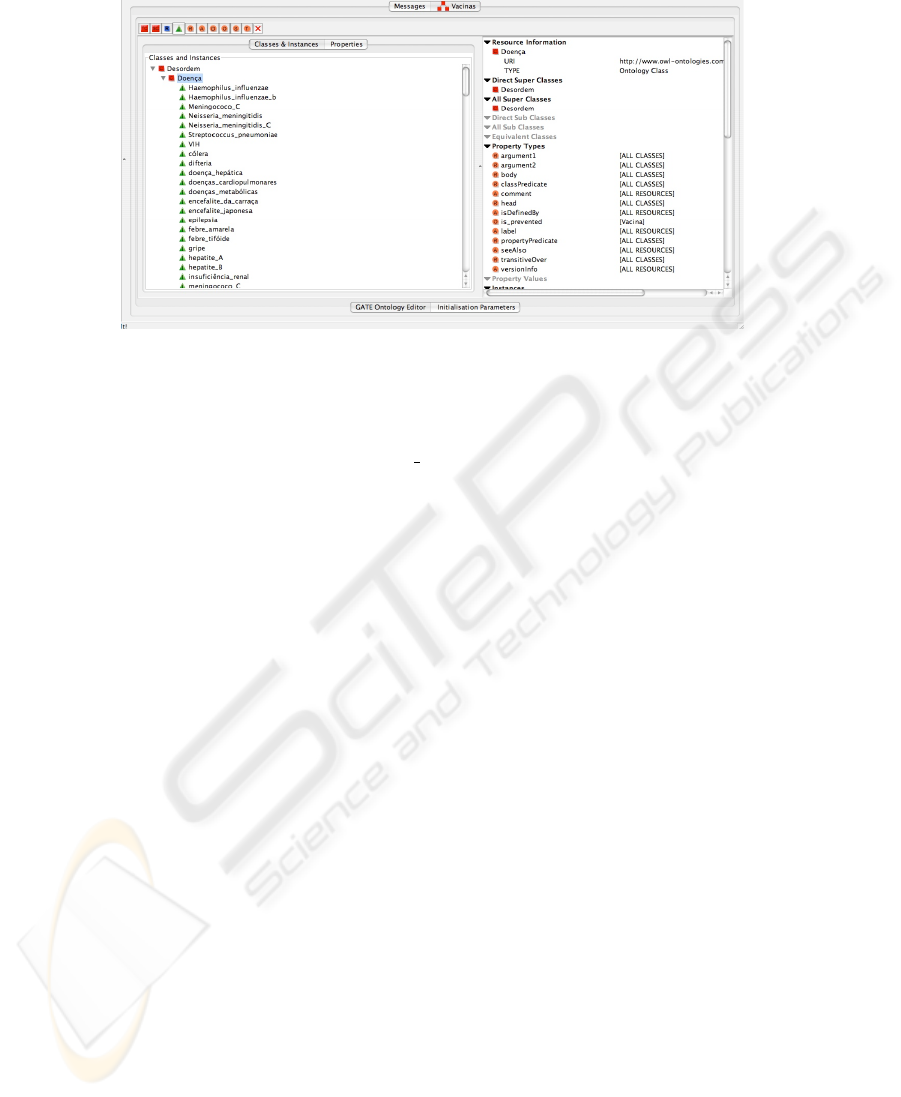
Fig. 3. Ontology Populated with vaccination information.
(80%)’ is found, it states that four out of five times that a disease individual (e.g. Tuber-
culosis) is mentioned it is followed by a vaccine individual (e.g. BCG). In this specific
case, this means the relationship BCG ’is against’ Tuberculosis can be inferred and the
RDF [9] triple can be automatically added.
The association mining algorithm algorithm [1] used to compute the association
rules can be stated as follows: Let I = {i
1
, ..., i
n
} be a set of items and D a set
of transactions (the entities). Each transaction consists of a subset of item in I. An
association rule is an implication of the form X → Y , where X ⊂ I, Y ⊂ I, and
X ∩ Y 6= ∅. The rule X → Y holds in D with confidence c if c% of annotations in
D that support X also support Y. The rule has support s in D if s% if transaction in D
contain X ∪ Y . The problem of mining association rules is to generate all association
rules in D that have a support and confidence greater than the user specified minimum
support and minimum confidence.
To mine frequent occurring entities we considered I as the set of all entities identi-
fied, I = {age, disease, acronym, bodypart, dosage, interaction, reaction, weight} and
D a list with the annotations made for each sentence of the PVP. An excerpt of the
database developed is in Table 2. Then we run the association rule miner, CBA [10],
which is based on the Apriori algorithm in [1]. This algorithm works in two steps. In
the first step, it finds all frequent pattern from a set of transactions that satisfy a user-
specified minimum support. In the second step, it generates rules from the discovered
frequent patterns, being in our case possible relationships between the instances anno-
tated in the PVP. In our work we defined a pattern as frequent if it appears in more than
20% (minimum support) of the annotated sentences.
100
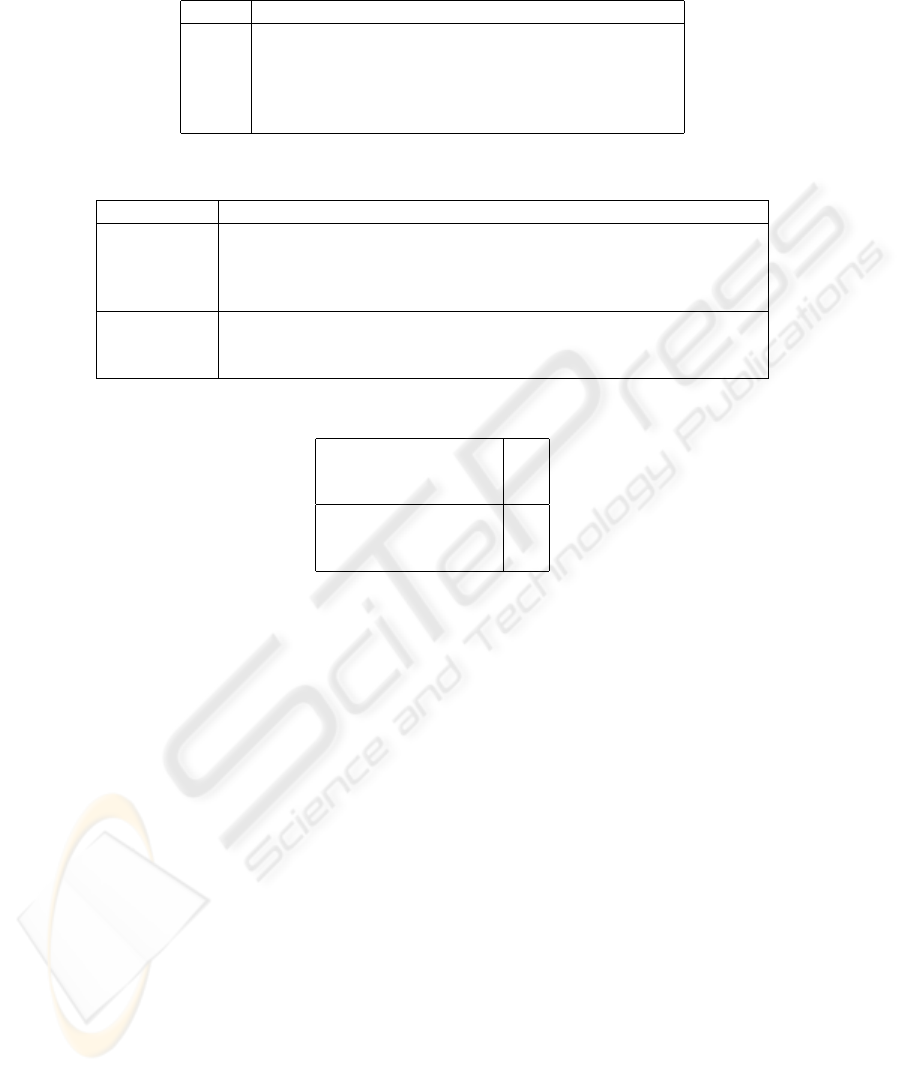
Table 2. An excerpt of the database D, sentences 100 to 104.
Sentence Annotation vectors
100 {age, acronym}
101 {age, age, weight}
102 {age, dosage, disease, disease, acronym, dosage, acronym}
103 {acronym, acronym}
104 {age, acronym, acronym}
Table 3. Results for Information Extraction task.
DISEASE ACRONYM AGE BODYPART DOSAGE INTERACTION REACTION WEIGHT
Correct matches 225 294 181 14 90 10 156 8
Partially correct 0 0 1 0 3 0 0 0
Missing 0 0 6 0 0 0 0 0
Total 225 294 188 14 93 10 156 8
Recall 1,00 1,00 0,96 1,00 0,97 1,00 1,00 1,00
Precision 1,00 1,00 1,00 1,00 1,00 1,00 1,00 1,00
F - measure 1,00 1,00 0,98 1,00 0,98 1,00 1,00 1,00
Table 4. Overall Information Extraction Results.
Correct Matches 978
Partially Correct matches 4
Missing 6
Recall 0,991
Precision 1,000
F - measure 0,995
5 Results
5.1 Information Extraction
Information extraction results are summarized in Table 3. The lines show the number
of entities correctly matched by the system, the ones partially correct and the number
of entities the system was not able to identify. Results in terms of recall, precision and
F-measure are in the bottom lines of the table. Table 3 shows results for each annotation
type, while Table 4 presents the overall results.
Precision of 100% was obtained for all entities annotated. This means that every
individual added in the ontology was correctly associated with the corresponding class.
Recall values indicate that all the information existing on the PVP was also added in
the ontology, except information concerning Age and Dosage properties of Vaccine for
which, respectively, 7 and 3 individuals are missing.
5.2 Relationships between Individuals
Association rule mining algorithm identified three association rules with a support of
20% and confidence of 80%. Figure 5.2 presents those rules. The first rule indicates
that 79.38% of the times that a disease is mentioned it is followed by an acronym of
101

a vaccine. Considering the area and the concrete aim of the PVP, it is possible to infer
that approximately four out of five times that a disease name is followed by a vaccine
acronym in a sentence the information is referring to a specific disease and the vaccine
that prevents it. The same analysis can be done for the second rule. In this case, we can
infer information about the vaccine that should be applied at a specific age, while the
third rule states that approximately 20% of the times information regarding the dosage
recommended for each vaccine is mentioned after the vaccine acronym in a sentence.
Number of rules = 3
(1) {DISEASE} -> {ACRONYM} 79.68%
(2) {AGE} -> {ACRONYM} 69.73%
(3) {ACRONYM} -> {DOSAGE} 20.83%
Fig. 4. Association mining rules.
6 Conclusions and Future Work
A system which identifies relevant entities on medical texts and automatically popu-
lates a vaccination ontology with new instances of classes is presented. The several
steps performed to model the vaccination ontology are described and a first attempt
to automatically add inter-instances relationships was performed resulting on two as-
sociation rules with a high level of confidence and suggests a third rule that could be
implemented after some further analysis.
The combination of the OWL populated ontology with SWRL rules provides clin-
ical decision support by introducing the possibility of offering suggestions when rules
are broken or exceptions made, or the suggestion of treatments based on patient con-
ditions. For instance, a children with two months has to be vaccinated against several
diseases. The system can suggest the appropriate vaccines to the physicians, reducing
consultation time and allowing increased safety and satisfaction.
The system can be extended to provide decision support on a deeper level. For in-
stance an active or a passive validation of medical reports can be done. The idea behind
the active validation of medical reports is to support an automatic and dynamic valida-
tion of decisions made over the content of the document by applying contextually rel-
evant rules to components of the documents during consultation. This is accomplished
by executing rules on semantic annotations and relationships to the ontology. A passive
validation can be done on documents referring to older consultations allowing the de-
tection of relationships between symptoms, patient details and treatments and enriching
the ontology with such information.
Even thought the ontology is already modeled and populated, the approach to au-
tomatize the inter-instance relationship population, needs further developed. Combining
the association rules and part-of-speech information to map between the inter-class re-
lationships describe in the guidelines and the ones defined on the ontology would give
further knowledge. These relationships are most of the times described by verbs. Using
available knowledge sources like wiki dictionaries one could be able to find relations
102

between the verbs used in the guidelines and the ones used in the relationship descrip-
tion, allowing an automatic mapping between concepts.
References
1. Agrawal, R., Srikant, R.: Fast algorithms for mining association rules. 20th International Con-
ference Very Large Data Bases, VLDB, 1215: 487-499, 1994
2. Cunha, J.P.C., Cruz, I., Oliveira, I., Pereira, A.S., Costa, C.T., Oliveira, A.M., Pereira, A.:
The RTS project: Promoting secure and effective clinical telematic communication within the
Aveiro region. In eHealth 2006 High Level Conference . Malaga, ES, Maio 2006 . p. 1-10
3. Cunningham, H., Maynard, D., Bontcheva, K. , Tablan, V., and Ursu, C.: The GATE User
Guide. 2002 http://gate.ac.uk/.
4. Cunningham, H., Maynard, D., Bontcheva, K. , Tablan, V.: GATE: A Framework and Graph-
ical Development Environment for Robust NLP Tools and Applications. Proceedings of
the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL’02).
Philadelphia, July 2002
5. Direc
˜
ao Geral da Sa
´
ude Programa Nacional de Vacina c
˜
ao 2006, Orienta
˜
oes T
´
ecnicas No 10
2006
6. Ferreira, L., Teixeira, A., Cunha, J.P.S.: Information Extraction from Medical Reports. In 3rd
International Workshop on Natural Language Understanding and Cognitive Science (NLUCS-
2006), Paphos, Cyprus, May 2006.
7. Herman, T.D., Liu, F., Sagaram, D., Raoul, K., Fontelo, P., Kohl, K., Payne, D.: Creating a
vaccine adverse event ontology for public health. AMIA Annu Symp Proc. 2005, 978
8. Horrocks, I, Patel-Schneider, P.F., Boley, H., Tabet, S, Grosof, B., Dean, M.: SWRL: A Se-
mantic Web Rule Language - Combining OWL and RuleML. W3C Member Submission,
http://www.w3.org/Submission/SWRL/, May 2004.
9. Lassila, O., Swick, R.R.: Resource Description Framework (RDF) Model and Syntax. W3C
Working Draft, World Wide Web Consortium, 1998; http://www.w3.org/TR/WD-rdf-syntax/.
10. Liu, B., Hsu, W., Ma, Y.: Integrating Classification and Association Rule Mining. KDD-98,
1998
11. McGuinness, D. L., Harmelen, F. , eds. OWL Web Ontology Language Overview. W3C
Proposed Recommendation, December 2003, http://www.w3.org/TR/owl-features/.
12. Sager, N., Lyman, M., Bucknall, C., Nhan, L.J., Tick, L.J.: Natural language processing and
the representation of clinical data. J. Am. Med. Informatics Assoc. March, 1994, 1(2): 142-60.
13. Serban, R., Teije, A., Harmelen, F., Marcos, M., Polo-Conde, C.: Extraction and use of lin-
guistic patterns for modelling medical guidelines. Artif. Intell. Med Journal, 2007, 39:137-149
14. Schr
¨
oder, M.: Knowledge-based processing of medical language: A language engineering
approach. Proceeding of GWAI’92, Bonn, September 1992
15. Sheth, A. P., Agrawal S., Lathem J., Oldham N., Wingate H., Yadav P., Gallagher K.: Active
Semantic Electronic Medical Record. International Semantic Web Conference 2006: 913–926
16. SNOMED Clinical Terms Guide. College of American Pathologists.
17. UMLS KNOWLEDGE SOURCES. 14th Edition. National Institutes of Health Department
of Health and Human Services. U.S. National Library of Medicine.
103
