
TOWARDS INTEROPERABILITY IN e-HEALTH SYSTEMS
A Three-Dimensional Approach based on Standards and Semantics
Jose Manuel Gómez-Pérez
1
, Sandra Kohler
1
, Ricardo Melero
1
, Pablo Serrano
2
, Leonardo Lezcano
3
Miguel Angel Sicilia
3
, Ana Iglesias
4
, Elena Castro
4
, Margarita Rubio
5
and Manuel de Buenaga
5
1
iSOCO S.A, Pedro de Valdivia 10, Madrid, Spain
2
Hospital de Fuenlabrada, Madrid, Spain
3
Universidad de Alcalá de Henares, Madrid, Spain
4
Universidad Carlos III, Madrid, Spain
5
Universidad Europea de Madrid, Madrid, Spain
Keywords: Semantic interoperability in eHealth, CEN 13606, Archetypes, NLP, OWL, SNOMED.
Abstract: The interoperability problem in eHealth can only be addressed by means of combining standards and
technology. However, these alone do not suffice. An appropriate framework that articulates such
combination is required. In this paper, we adopt a three-dimensional (information, concept, and inference)
approach for such framework, based on OWL as formal language for terminological and ontological health
resources, SNOMED CT as lexical backbone for all such resources, and the standard CEN 13606 for
representing EHRs. Based on such framework, we propose a novel form for creating and supporting
networks of clinical terminologies. Additionally, we propose a number of software modules to semantically
process and exploit EHRs, including NLP-based search and inference, which can support medical
applications in heterogeneous and distributed eHealth systems.
1 INTRODUCTION
Healthcare is one of the most information-intensive
sectors of European economies, expected to greatly
profit from research in information and
communication technologies. However, the general
feeling is that, to date, health information
technologies have been mostly the realm of
enthusiasts and the computer wave has not yet
completely arrived.
The European eHealth Action Plan
1
provides a
mid-term roadmap for improvement of the Health
sector. One of the most challenging issues identified
addresses the interoperability problem between
different e-health systems. Such problem is partially
due to the exponential increase of the number of
medical terminologies (SNOMED CT
2
, MedDra
3
,
1
ec.europa.eu/information_society/activities/health/policy
2
www.snomed.org
3
www.meddramsso.com/MSSOWeb/index.htm
etc.), ontologies (GALEN
4
, FMA
5
, etc), and
classifications of diseases and related medical events
and concepts (ICD
6
, CPT
7
, etc.) that eventually need
to interoperate with one and another. The same
report highlights the need for standardization as the
key piece to ensure interoperability in this roadmap.
Making eHealth systems interoperable by means of
consensual, standard data formats and protocols will
allow for a significant step forward towards
satisfactory healthcare, accomplishing a number of
goals like improvement of the quality of patient care,
reduction of medical errors, and therefore savings in
terms both of human and economic costs.
Experiences on semi-automated local health systems
have shown a lack of underlying standards for data
exchange, emphasizing as a result that the gap
4
www.open-galen.com/index.html
5
sig.biostr.washington.edu/projects/fm/index.html
6
www.cdc.gov/nchs/icd9.htm
7
www.ama-assn.org/ama/pub/category/3113.html
205
Gómez-Pérez J., Kohler S., Melero R., Serrano P., Lezcano L., Sicilia M., Iglesias A., Castro E., Rubio M. and De Buenaga M. (2009).
TOWARDS INTEROPERABILITY IN e-HEALTH SYSTEMS - A Three-Dimensional Approach based on Standards and Semantics.
In Proceedings of the International Conference on Health Informatics, pages 205-210
DOI: 10.5220/0001539302050210
Copyright
c
SciTePress
between consumer expectations and actual service
delivery remains unabridged.
The roadmap towards leveraging the
interoperability problem in eHealth has been
favoured by significant advances in information
technologies, especially with respect to knowledge
representation and interoperability. Particularly,
large effort has been invested on the field of
semantic technologies, finally coming of age and
entering the plateau of commercial productivity. The
usage of ontologies
8
as the main asset of semantic
technologies is currently supported by a wide range
of tools for ontology construction, storing, feeding,
evolution and evaluation like Protégé
9
or the NeOn
Toolkit
10
. In addition, mature methodologies and
standards allow for accurate scheduling of ontology
and application developments in terms of effort,
time, quality and finance resources.
As a consequence, the Semantic Web initiative
has proved to offer a reliable solution for large scale
integration and representation problems, which can
significantly contribute to alleviating the
interoperability problem in the Health domain. In
parallel, standardization efforts are going on in
various subdomains for electronic interchange of
clinical, financial, and administrative information
among health care oriented computer systems, for
e.g. definition of communication standards (HL7
11
)
or information models for electronic health record
(ISO/CEN 13606
12
norm and openEHR
13
specifications). The work presented in this paper
combines both approaches (uptake of standards and
semantic technologies) to tackle the interoperability
problem in the Health domain.
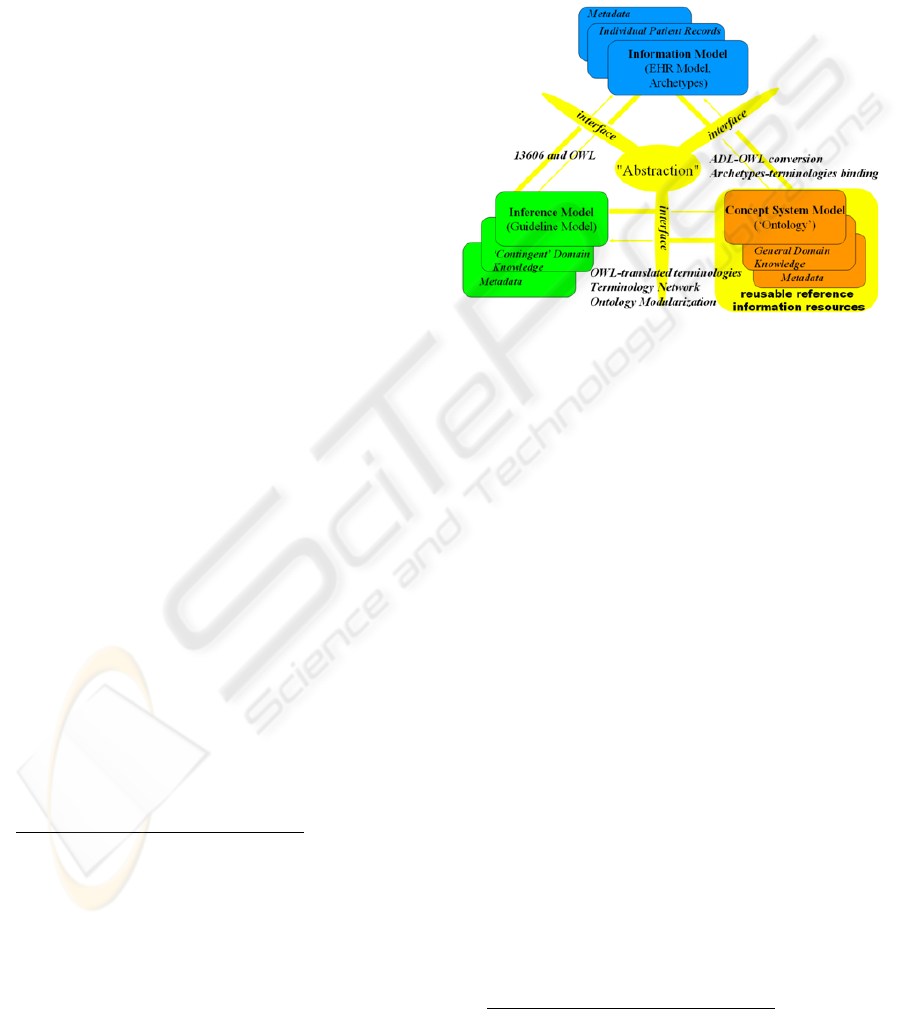
Complete information Health systems need to
address three main dimensions (information, concept
system, and inference)
14
, as well as their associated
resources, which are developed independently. Each
of these components comprises the model itself,
knowledge about a given view of the domain,
metadata, and interfaces to the other components.
Figure 1 shows a version of this vision, instantiated
with the solutions adopted in our approach, based on
OWL
15
as formal language for terminological and
ontological health resources, SNOMED CT as
lexical backbone for all such resources, and the
8
en.wikipedia.org/wiki/Ontology_(computer_science)
9
http://protege.stanford.edu/
10
www.neon-toolkit.org
11
www.hl7.org
12
www.chime.ucl.ac.uk/resources/CEN/EN13606-1/N06-
02_prEN13606-1_20060209.pdf
13
www.openehr.org
14
www.semantichealth.org
15
www.w3.org/TR/owl-ref
standard CEN 13606 (together with the ADL
16
language for archetype definition). The information
dimension deals with high quality, structured and
timely data collection and representation, allowing
building an information framework for electronic
health records (EHRs). In the present work, we
exploit archetypes as formalism for modelling the
required structures for EHR and defining the context
of the clinical domain where such records belong.
Figure 1: Components of a complete Health system
(source: semantichealth.org).
In our approach, Natural Language Processing
(NLP) support for analyzing patient records is part
of the information dimension and uses the
terminologies provided by the terminology server in
the concept system dimension to identify and
process the information contained in the records. On
the other hand, we use NLP (Jurafsky & Martin,
2000) for extracting data and information from EHR
(free text documents) for further processing. The
NLP of the EHR uses the terminologies provided by
the terminology server to identify and process the
information contained in the records and enable
inferences using the EHR.
The information dimension lies on top of the
concept system dimension, which deals with all the
available terminological and ontological resources
and provides the other two dimensions with uniform
access to such resources. We have addressed the
problem of managing all these resources through the
development of a terminology server, which
consequently allows relating them in a terminology
network.
Finally, the inference dimension exploits both
the concept system and information dimension to
discover new knowledge. Inference and semantic
search through NLP interfaces are some of the most
16
www.openehr.org/drafts/ADL-12draftF.pdf
HEALTHINF 2009 - International Conference on Health Informatics
206

immediate functionalities on top of which
applications of this vision can be implemented.
The remainder of this paper is structured as
follows: Sections 2 provides an insight to the three
layers of the model, section 3 illustrates the
integrated use of the described infrastructure and
section 4 concludes the paper by describing ongoing
work.
2 THREE LAYER MODEL
2.1 Information Dimension
As introduced above, the information dimension
deals with representing and structuring EHRs. In this
work, we approach this dimension from three
different and complementary perspectives:
1.) The aim of the ISO/CEN 13606 standard is to
normalize the transfer of information between EHRs
systems in an interoperable fashion, without
specifying how to implement them. The reference
model represents the general features of the
components of the health record, how they are
organized and the context information needed to
satisfy both the ethical and legal requirements of the
record. This model defines building blocks for a
formal representation of EHRs. An archetype is the
definition of a hierarchical combination of
components of the reference model, which restrict it
(giving the names, possible types of data, default
values, cardinality, etc.), to model clinical concepts
of the knowledge domain. These structures, although
sufficiently stable, may be modified or substituted
by others as clinical practice evolves.
The Archetype Definition Language (ADL)
allows expressing archetypes. An archetype starts
with a header section followed by a definition
section and an ontology section. The header includes
a unique identifier for the archetype, a code
identifying the clinical concept defined by the
archetype. The definition section contains the
restrictions in a tree-like structure created from the
reference information model. This structure
constrains the cardinality and content of the
information model instances compliant with the
archetype. Codes representing the meanings of
nodes and constraints on text or terms as well as
bindings to terminologies such as SNOMED, are
stated in the ontology section of an archetype.
2.) Processing EHRs requires handling structured,
semi-structured and unstructured data. To process
unstructured data, which is generally free text such
as clinical notes taken by doctors and nursing staff
during patient visits, tools are needed that work
automatically with the language and allow
information to be extracted so it can be easily stored
and consulted. This is especially important in
processes related to Patient Safety.
A series of additional problems exist for working
with reports in free text written by clinical
personnel: heavy use of acronyms and abbreviations;
spelling errors; and including information on people
or organizations, which must be anonymized in
order to comply with laws on health information.
Furthermore, most of the works in this area focus on
the English language, where specific resources in
biomedicine can be found, as MESH, UMLS
(Bodenreider 2004), etc. Nevertheless, in the case of
other languages, such as Spanish, relevant studies
dealing with hospital reports or clinical notes have
not been carried out yet.
Herein we present MOSTAS: A MOrpho-
Semantic Tagger, Anonymizer and Spellchecker for
Biomedical Texts, in order to address these
problems in the information dimension of eHealth
systems. The main objective of MOSTAS is to
analyze clinical reports in Spanish using the
ontological and lexical resources available for the
Spanish language in order to first, pre-process the
clinical reports so that they can be anonymized,
abbreviations and acronyms can be detected and
expanded, medical concepts in the application
domain can be detected. The system’s output is an
XML document with morpho-semantic information
that will facilitate later information retrieval of these
texts.
3.) The remaining module in the information
dimension of our system deals with semantic search
through NLP interfaces using conceptual
knowledge. This module is oriented to implement
main features of document indexing based on the
exploitation of knowledge and ontological resources
included in an integrated way in UMLS, as
SNOMED and MeSH. For the design and evaluation
of the NLP semantic search module, we have
developed a basic system offering interconnection
between health records and a set of scientific
information and health news. Given a query in
submitted by a person, it first retrieves a list of
medical records ordered by relevance in three steps:
i) the query is expanded using concepts included in a
biomedical ontology (i.e.: UMLS); ii) medical
records are ranked using a representation based on
biomedical concepts; iii) then, the user can choose a
record and the system will retrieve several lists of
ranked documents in English: from Pubmed news,
or from article abstracts.
TOWARDS INTEROPERABILITY IN e-HEALTH SYSTEMS - A Three-Dimensional Approach based on Standards and
Semantics
207
2.2 Concept System Model
1.) Addressing the concept system dimension
requires devising a way to deal with the
interoperability problem between the available
terminologies and ontological resources used by the
different computer health systems. In our
framework, we have approached this problem by
developing a terminology server, an open platform
for: 1) Normalizing pre-existing terminologies as
OWL ontologies, 2) Importing available ontological
resources into the server, 3) Relating all these
resources with each other in a terminology network,
where equivalent terms are connected, and 4)
Visualizing and browsing such network. By open,
we mean that the terminology server is fully
extendable with new terminologies, which can be
plugged in as desired. Currently the following
terminologies have been integrated into the system:
ICD-9, official classification of the WHO
17
for
diseases and health problems, CPC-2
18
, international
classification for primary care, SNOMED CT, the
most extensive terminology in medical
terminologies, Local Terminologies, containing
terms used by hospital clinic personnel in their
patient records and notes. Using OWL (the W3C
standard ontology language) for representing
normalized medical terminologies is accompanied
by a large number of advantages. In a nutshell, these
can be summarized as: high expressivity, reasoning
capabilities, inference, and a wide tool and
infrastructure support favoured by its status as a
standard and the ongoing contributions from the
knowledge engineering community. Additionally the
number of medical resources available in OWL is
dramatically increasing. As a consequence, in this
work, we have adopted OWL as the reference
language for medical terminology. As part of our
approach, it was necessary to translate legacy
terminologies into OWL in order to incorporate
them into the terminology server. The most relevant
case is SNOMED CT, which required a specific
treatment due to its size and complexity. The
SNOMED CT terminology is distributed across
three text files: one containing the English terms
(>300,000), another for term names in other
languages (Spanish in our version) plus their
corresponding preferred term and synonyms, and a
third one describing the SNOMED CT taxonomy
and the relations between the terms (>1.000,000).
In the translation process, we neither tried to
17
www.who.int
18
www.globalfamilydoctor.com/wicc/pagers/english.pdf
improve the overall quality of SNOMED CT
(though several errors were detected (Schulz et al,
2007), (Rector, 2007)) nor did we modify the
concept names. The resulting OWL file contains the
most relevant parts of the terminology without
extending its semantics.
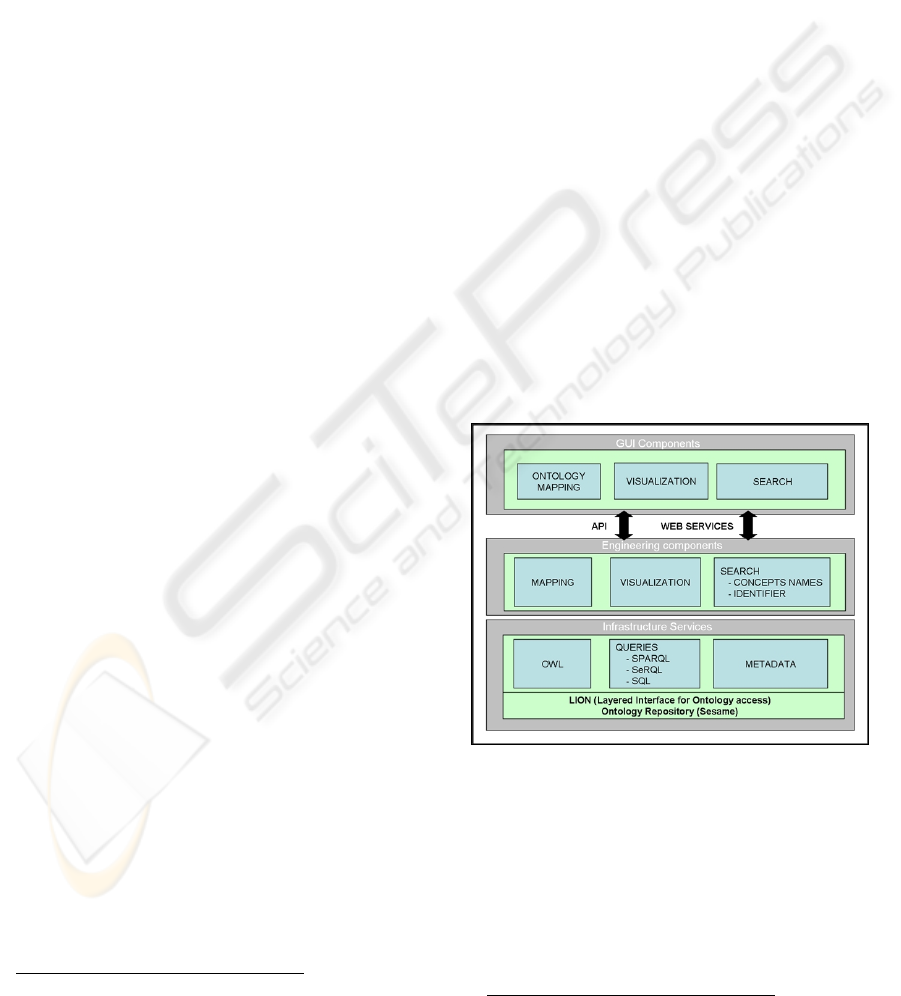
2.) The terminology server follows a three layer
architecture (Figure 2). The lower level stores,
maintains and provides access to the terminologies,
in their OWL forms, currently stored in a Sesame
repository
19
. The same level allows managing
metadata about the terminologies, like e.g. the
terminology subdomain, the authoring institution, a
short description, etc, which can be useful during
search. The middle layer contains engineering
components that implement three basic
functionalities on top of the lower layer: i) search of
terms both in a single terminology or across several,
ii) mapping related terms of different terminologies,
and iii) term visualization and browsing in and
across terminologies. The higher level of the
architecture contains the GUI components of the
user applications exploiting the functionalities
provided by the underlying layers of the terminology
server. The GUI components can access such
functionalities either programmatically, via a Java
API, or in a loosely coupled way, through web
services.
Figure 2: Terminology server three layer architecture.
3.) Currently, the terminology server provides two
different types of search: The identifier-based
search, where the user types the code of the term in a
before specified terminology and the name-based
search, where the user types the name of the concept
or a part of it. As a shared characteristic, both
searches return the sought term (if any) and situate it
in the terminology network, showing the terms
19
www.openrdf.org
HEALTHINF 2009 - International Conference on Health Informatics
208

related with it in the other terminologies. This search
scenario requires the definition of a terminology
network, where the different terminologies are
connected by means of related terms. For example,
“Acne disorder” in SNOMED CT corresponds to
“Acne” in ICD-9. Currently, in order to support
users in defining the mappings of a term against the
corresponding terms of the remaining terminologies
of the network, we first search its name in those
terminologies. Then, we use SNOMED CT as a
terminology gateway in case no such term name is
detected, i.e. we automatically fetch its synonyms
from SNOMED CT and then search the synonyms
instead. If still no solution is found, the terminology
server tokenizes the term name, discarding stop-
words and starts a new search with these tokens and
their synonyms. Future work includes using
ontology matching
20
techniques that exploit the
semantics of the terminologies in OWL.
2.3 Inference Model
This section broadly reports on an approach to
convert ADL definitions to OWL and then attach
rules to the semantic version of the archetypes.
Let us consider the following situation. A health
care information system that receives some
OBSERVATION (e.g. “blood pressure”) entry (no
matter where from but fulfilling an archetype
specification) is able to syntactically understand
such information and therefore may deliver it to a
professional, who proceeds with the
OBSERVATION assessment. This is clearly a great
advance for the interoperability of medical systems
but it would be even more interesting if the
observation archetype could tell not only how to
manipulate observation’s values but also how to
assess and evaluate them. Every task that depends on
data analysis and conclusion arrival usually requires
the presence of an expert with enough knowledge to
make a good decision. However if we separate tacit
from explicit knowledge then we could add the latter
in the archetyped concept so the expert only needed
to deal with the former one.
Unfortunately, the ADL language does not
provide support for rules and inference which are
important pieces of clinical knowledge. Besides,
while one of the greatest advantages of two-level
modelling (Beale, 2002) is the carrying out of
archetype definition as a decentralized process, it
allows for contradictory viewpoints to coexist or
even false information to be provided. In addition, a
higher level of normalization of clinical knowledge
could be achieved, encouraging for automated
20
www.ontologymatching.org
means to reuse knowledge expressed in the form of
rules, which follows the same philosophy of sharing
archetypes.
SWRL
21
is a W3C recommendation developed to
improve OWL limitations, in terms of inference, by
means of rules. In combination, they add
considerable expressive power to the Semantic Web.
Furthermore, by merging SWRL rules with OWL
ontologies, we will be able to partially automate
decision making process.
Concretely, the complete knowledge workflow,
from archetypes to inference, can be summarised as
follows: 1) Translating ADL to OWL, 2) Mapping
clinical data to OWL instances, 3) Adding SWRL
rules to the ontology, 4) Executing inference.
When translation is finished, the obtained
ontology file should be filled with instances of
concrete clinical data. Depending on the nature of
the data source, an adequate access approach should
be chosen to correctly map each field to individuals’
properties. From our perspective, preferred source
will be the one where supplied XML files are
compatible to the Reference Model syntax. In this
case, instance mapping is a straightforward process.
As a particular implementation, here we adopt an
inference process based on the Jess-Java bridge
provided with the Protégé ontology editor
(Golbreich and Imai, 2004). The Protégé SWRL
Editor is an extension to Protégé-OWL that permits
interactive editing of SWRL rules. It generates OWL
files that include attached SWRL expressions.
The resulting OWL file, enriched with inferred
knowledge, has many possible destinations. For
example it can be directly delivered to the end user
through a compatible interface or stored in a
repository. In the clinical domain, these results
provide means for automatically improving decision
making and monitoring tasks.
3 INTEGRATION
The following example, focused on preventing
pressure ulcers in hospitalized patients illustrates an
integrated, practical use of the three-dimensional
(Information, Concept, and Inference) architecture
described in this paper. Pressure ulcers are a severe
problem for bedridden patients caused by many
different reasons like friction or humidity, which,
not treated in time can become live-threatening. The
goal of the hypothetical system described in this
example is to automatically produce an alarm if a
risk of ulcer is detected for any patient.
21
www.w3.org/Submission/SWRL
TOWARDS INTEROPERABILITY IN e-HEALTH SYSTEMS - A Three-Dimensional Approach based on Standards and
Semantics
209
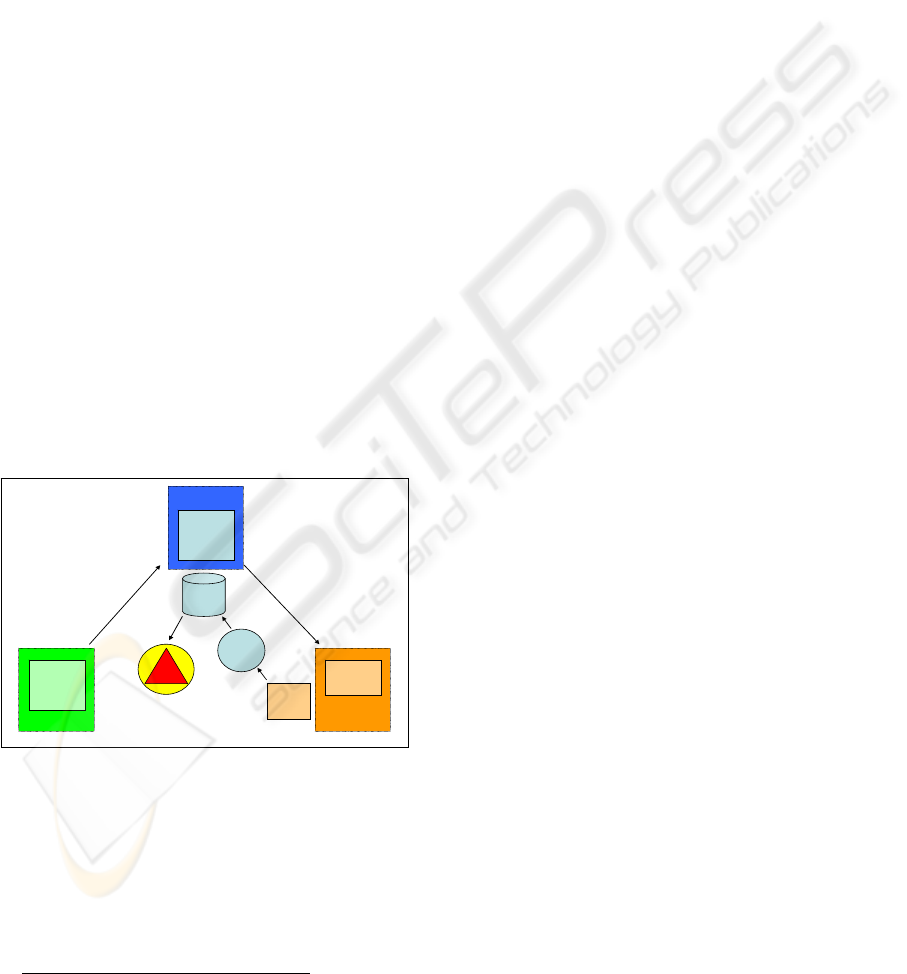
First, we define an archetype in the CEN 13606
standard (Information Model) by using the
knowledge of the clinical personnel of this concrete
domain (Inference Model), see Figure 3. The
archetype contains all the information related to the
field of pressure ulcers and defines rules for
identifying possible risks (for example: If the patient
is not able to leave the bed, the risk increases
22
). The
next step is the linking of the CEN standard with a
reference terminology, in our case SNOMED CT
(Concept System Model).
Now, the risk-detecting protocol can start: The
system is periodically fed with information about the
patients, contained in clinical databases. The terms
contained in such information are processed by the
terminology server, expanded using the mappings
defined across the available terminology network.
This allows the NLP systems to detect occurrences
of the words in the EHRs, which are also corrected if
they had been previously misspelled or abbreviated.
With the information found in the EHRs the rules
built as an extension of the OWL ontologies
corresponding to the ADLs resulting from
implementing the archetype, are immediately
triggered and, if a risk of pressure ulcer is detected
the alarm is triggered.
Additionally, semantic search may offer
information from different EHRs and scientific or
reference documentation related with the particular
case on hand, facilitating decision making to the
clinical personnel.
Figure 3: Example for the interactivity.
4 CONCLUSIONS
AND ONGOING WORK
The interoperability problem in eHealth can only be
addressed by means of combining standards and
22
http://www.sanitariascaligera.com/index.php?option=com_cont
ent&task=view&id=44&Itemid=69&lang=en
technology. An appropriate framework that
articulates such combination is required. In this
paper, we adopt a three-dimensional (information,
concept, and inference) approach for such
framework. Based on this framework, we have
proposed a novel form of relating the different
terminologies with each other by means of a
terminology server that supports a clinical
terminology network. On top of that, we have also
proposed a number of modules to semantically
process and exploit EHRs, including NLP-based
search and inference, which support applications like
e.g. automatic detection of pressure ulcers.
Nevertheless, all this work is still preliminary,
and we are addressing further tests and evaluation in
real-world systems. Ongoing work lies in this
direction, aiming to demonstrate our approach for
e.g. personal health records. Furthermore, we will
continue the integration of semantic technology in
this framework, especially in the concept dimension,
incorporating novel ontology modularization,
mapping, and context technology in order to
facilitate management of complex and large
terminologies as in the case of SNOMED CT.
ACKNOWLEDGEMENTS
This work has been funded as part of the Spanish
nationally funded projects ISSE (FIT-350300-2007-
75) and CISEP (FIT-350301-2007-18). We also
acknowledge IST-2005-027595 EU project NeOn.
REFERENCES
Jurafsky D. and Martin, J.H., 2000. Speech and language
processing: An Introduction to Natural Language
Processing, Computational Linguistics, and Speech
Recognition. Prentice-Hall.
Beale T., 2002. Archetypes: Constraint-based Domain
Models for Future - proof Information Systems.
OOPSLA, workshop on behavioural semantics.
Available at http://www.deepthought.com.au.
Golbreich, C. and Imai, A., 2004. Combining SWRL rules
and OWL ontologies with Protégé OWL Plugin, Jess,
and Racer. 7th International Protégé Conference,
Bethesda, MD
Bodenreider O., 2004. The Unified Medical Language
System (UMLS): integrating biomedical terminology.
Nucleic Acids Research 2004, 32:D267-D270
Schulz, S., Suntisrivaraporn, B., Baader, F. 2007.
SNOMED CT’s Problem List: Ontologists’ and
Logicians’ Therapy Suggestions, Medinfo.
Rector, A., 2007. What's in a Code?: Towards a Formal
Account of the Relation of Ontologies and Coding
Systems, Medinfo
Archetypes
Domain
Knowledge
Snomed
ICD-9, ICPC-2
etc.
Clinical personnel
develops
Archeype
In CEN-
standard
Linking CEN with
terminologies
EHR
repository
Concept System
Model
Inference Model
Information
Model
Term.
Server
ALARM
Applying
rules of
archetype
START
OWL
NLP
HEALTHINF 2009 - International Conference on Health Informatics
210
