
CONTEXT-DRIVEN ONTOLOGICAL
ANNOTATIONS IN DICOM IMAGES
Towards Semantic PACS
Manuel Möller
German Research Center for Artificial Intelligence, Kaiserslautern, Germany
Saikat Mukherjee
Siemens Corporate Research, Princeton, New Jersey, U.S.A.
Keywords:
Semantic Search, Medical Image Retrieval, Semantic PACS.
Abstract:
The enormous volume of medical images and the complexity of clinical information systems make searching
for relevant images a challenging task. We describe techniques for annotating and searching medical images
using ontological semantic concepts. In contrast to extant multimedia semantic annotation work, our technique
uses the context from mappings between multiple ontologies to constrain the semantic space and quickly
identify relevant concepts. We have implemented a system using the FMA and RadLex anatomical ontologies,
the ICD disease taxonomy, and have coupled the techniques with the DICOM standard for easy deployment
in current PAC environments. Preliminary quantitative and qualitative experiments validate the effectiveness
of the techniques.
1 INTRODUCTION
Advances in medical imaging have enormously in-
creased the volume of images produced in clinical fa-
cilities. At the same time, modern hospital informa-
tion systems have also become more complex. To-
day’s clinical facilities typically contain hospital in-
formation systems (HIS) for storing patient billing
and accounting information, radiological informa-
tion systems (RIS) for storing radiological reports,
and picture archiving and control systems (PACS) for
archiving medical images. Standardized languages
such as DICOM (Digital Imaging and Communica-
tion, http://medical.nema.org/) have been developed
for digitally representing the acquired images from
the various modalities. Apart from the image pixels, a
DICOM image also contains a header, which is used
to store certain patient information such as name, gen-
der, demographics, etc.
It has become challenging for clinicians to query
for and retrieve relevant historical data due to the vol-
ume of information as well as the complexity of infor-
mation systems. In particular, historical patient im-
ages are useful for analyzing images of a current ex-
amination since they help in understanding any pro-
gression of pathologies or development of recent ab-
normalities. In current PACS, radiologists can query
for historical images using only meta attributes stored
in the DICOM headers of the images. However,
these attributes do not contain any information about
the anatomy or disease associated with the image.
Hence, radiologists are often overwhelmed with irrel-
evant images not connected with the current exami-
nation. With current querying based only on patient
attributes, it is difficult for the radiologist to easily
search for images using semantic information such as
anatomy and disease.
In this paper, we describe semantic techniques
for searching medical images. Our approach re-
lies on annotating images with formal concepts
such that the images themselves become queriable.
Recent advances in the Semantic Web community
have made it possible for knowledge to be for-
malized in languages such as Web Ontology Lan-
guage OWL (McGuinness and van Harmelen, 2004),
data to be annotated in RDF Resource Descrip-
tion Format (Hayes, 2004). At the same time,
work on formalizing clinical knowledge has resulted
in vocabularies and ontologies such as Radiolo-
gist Lexicon (RadLex, http://www.radlex.org), ICD9
(http://www.cdc.gov/nchs/icd9.htm) and the Founda-
tional Model of Anatomy (FMA) (Rosse and Mejino,
294
Möller M. and Mukherjee S. (2009).
CONTEXT-DRIVEN ONTOLOGICAL ANNOTATIONS IN DICOM IMAGES - Towards Semantic PACS.
In Proceedings of the International Conference on Health Informatics, pages 294-299
DOI: 10.5220/0001550202940299
Copyright
c
SciTePress

2003). Our technique leverages on the work in Se-
mantic Web and clinical terminologies to annotate
and search medical images with semantic concepts
from two dimensions – anatomy and disease. We have
used RadLex and FMA as the knowledge sources for
anatomy and the ICD9 classification system for dis-
ease knowledge. The focus of our work has been on
using existing ontologies for defining the semantics
rather than creating a custom ontology from scratch.
Hence, recent works on extending RadLex to a radi-
ological ontology either in the RadiO initiative
1
or in
(Rubin, 2007) are complementary to our approach.
Existing techniques on multimedia semantic an-
notation often do not translate easily to medical im-
ages because clinicians have limited time to perform
the annotation. This becomes even more challenging
while annotating images with concepts from multi-
ple dimensions. Automated image annotation, while
having the ability to free up the clinician’s time, are
not yet scalable enough to be applied to arbitrary
anatomical regions and diseases. In our work, we
have created mappings between different ontologies
such that given concepts in one dimension we can use
the mapping context and quickly identify the relevant
concepts in other dimensions. Specifically, we have
devised automated techniques for mapping RadLex,
FMA and ICD9 taxonomies such that this context can
be automatically generated.
The work in (Rubin et al., 2008) is similar to our
research in annotating medical images with ontology-
based semantics and the use of context for faster an-
notation. However, their notion of context is not tied
to diseases but rather more anatomical. Since the rich
information in medical images is implicitly related to
diseases, it is important to capture this dimension as
part of the context.
Another limitation of easily using current multi-
media semantic annotation techniques for querying
medical images is that they are often not adapted to
today’s clinical standards and health information sys-
tems such as, in particular, PACS. In our approach,
the semantic annotations are directly included in the
headers of DICOM images and do not require any ad-
ditional storage mechanisms. A fast index of con-
cepts is looked up for results during query time and
the headers of relevant DICOM images are parsed to
retrieve the original annotations. Consequently, only
minimal infrastructural changes are required to con-
vert today’s PACS to tomorrow’s semantic PACS us-
ing our technique.
1
The Ontology of the Radiographic Image: From
RadLex to RadiO, IFOMIS, Saarland University,
http://www.bioontology.org/wiki/images/a/a5/Radiology1.
ppt
Note that the RIS contains additional information
beyond the basic patient attributes in DICOM head-
ers in today’s PACS. While in principle it is possible
to query the RIS for additional meta information, in
practice however, this becomes challenging due to the
unstructured text format of radiology reports as well
as due to the loose coupling between RIS and PACS
which makes seamless querying across both of them
together difficult. In contrast, our technique outlines
a different paradigm where images themselves can be
semantically enriched and queried.
The main contributions of our work are:
• Our technique enables medical images to be an-
notated with semantic concepts from anatomy and
disease ontologies, storing the annotations in the
headers of DICOM images, and querying the an-
notations using semantic concepts.
• We have integrated semantics from two dimen-
sions, anatomy and disease, using the Radlex,
FMA and ICD9. The integration is based upon
mapping between the anatomy and disease on-
tologies.
• We have used the integrated semantics to contex-
tualize the space of concepts in one dimension,
anatomy for instance, given concepts in the other
dimension, disease in this example, for faster
identification of relevant images.
The research described in this paper on seman-
tic annotation and retrieval is part of a broader effort
in semantic understanding of medical images in the
THESEUS MEDICO (Möller et al., 2008) project.
2 RELATED WORK
To the best of our knowledge, using formalized on-
tologies for enriching existing PACS has not been
extensively researched in the community. The work
in (Kahn et al., 2006) uses ontologies for integrating
PACS with other clinical data sources, such as RIS
and HIS, but does not address enriching medical im-
ages with additional semantics.
(Buitelaar et al., 2006) investigate methods for an-
notating multimedia data of all forms within one sin-
gle retrieval framework and could be used for inte-
grating heterogeneous clinical content from images to
text to genomics data.
There has also been sizable research in retriev-
ing images using ontology-based semantics as well
as machine learning (Carneiro et al., 2007). The
learning-based works, while being more scalable and
automated, use subjective semantics that does not for-
malize notions of domain specific concepts and terms.
CONTEXT-DRIVEN ONTOLOGICAL ANNOTATIONS IN DICOM IMAGES - Towards Semantic PACS
295
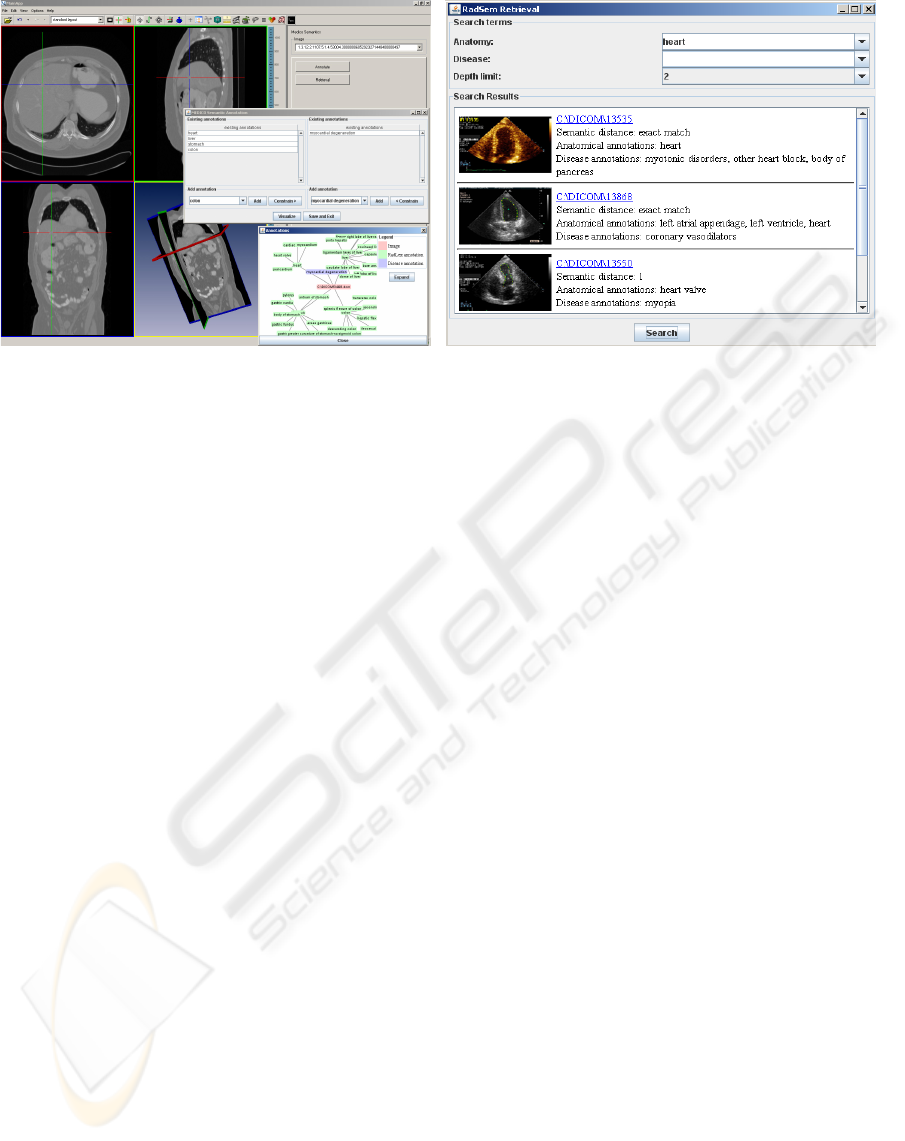
(a) (b)
Figure 1: Implementation of (a) Semantic Annotation, and (b) Semantic Retrieval within a 3D image browser.
A number of research publications in the area of
ontology-based image retrieval emphasize the neces-
sity to fuse sub-symbolic object recognition and ab-
stract domain knowledge. (Su et al., 2002) present a
system that aims at applying a knowledge-based ap-
proach to interpret X-ray images of bones and to iden-
tify the fractured regions.
3 SEMANTIC ANNOTATION AND
RETRIEVAL
The essence of our technique lies in annotating im-
ages with concepts from anatomy and disease ontolo-
gies. Subsequently, these images can be queried using
terms from these ontologies. We mapped the anatom-
ical semantics to disease such that annotations can
be performed efficiently and relevant images retrieved
with minimal query terms and greater flexibility.
We use RadLex as the central terminology of
anatomical concepts. RadLex contains a hierarchy of
anatomical concepts corresponding to entities which
can be identified in medical images. We also use
the FMA, with around 70, 000 concepts, as a second
source of formal human anatomical knowledge. We
wanted to use the rich set of concepts in the FMA, but
did not want to overload clinicians in using the FMA
for image annotation since our goal is to identify
anatomical landmarks. Furthermore, we use ICD9 as
the ontology of diseases. By parsing the main table
of ICD9 categories we constructed an OWL ontology
which reflects the hierarchical structure of the original
categorization within subClassOf relationships.
We have implemented simplistic heuristics for
mapping RadLex to FMA as well as mapping RadLex
to ICD9. This helps in constraining the space of con-
cepts in one dimension (e.g., disease) knowing pos-
sible concepts in the other dimension (e.g. anatomy)
and is an effective way to cope with 8, 389 RadLex
and 8, 686 ICD9 concepts.
Our RadLex FMA mapping technique identifies
correspondences between the pair of vocabularies by
comparing terms at the lexical level and checking
for perfect matches. This resulted in identifying
1, 259 identical concept names between the two data
sets. The ICD9-RadLex mapping uses information
in ICD9 which provides links between ICD9 iden-
tifiers and anatomical concepts. Our mapping tech-
nique was able to associate 3, 694 ICD9 terms to
RadLex concepts covering 42.5% of the ICD9 vo-
cabulary. While there are far more sophisticated on-
tology mapping techniques, we had to rely on rather
simple methods based on string comparison of con-
cept names due to scalability issues. For instance,
PhaseLib (http://phaselibs.opendfki.de), an ontology-
structure driven mapping technique was not able to
cope with FMA and RadLex even with 2GB of mem-
ory. We plan to improve our string matching tech-
niques for more complete mapping.
Fig. 2 gives an example of some of the relations
between the ontologies described above. For this
illustration we chose the anatomical concept “Heart”.
The three big boxes in light gray denote the three
ontologies used in the backend of RadSem. The
smaller boxes in dark gray represent concepts from
these ontologies, each with the identifier in the first
line of the box. From the RadLex ontology in OWL
we get only plain subClassOf relationships. These
relationships provide information, that there is a
connection, eg between “Heart” and “Pericardium”
HEALTHINF 2009 - International Conference on Health Informatics
296
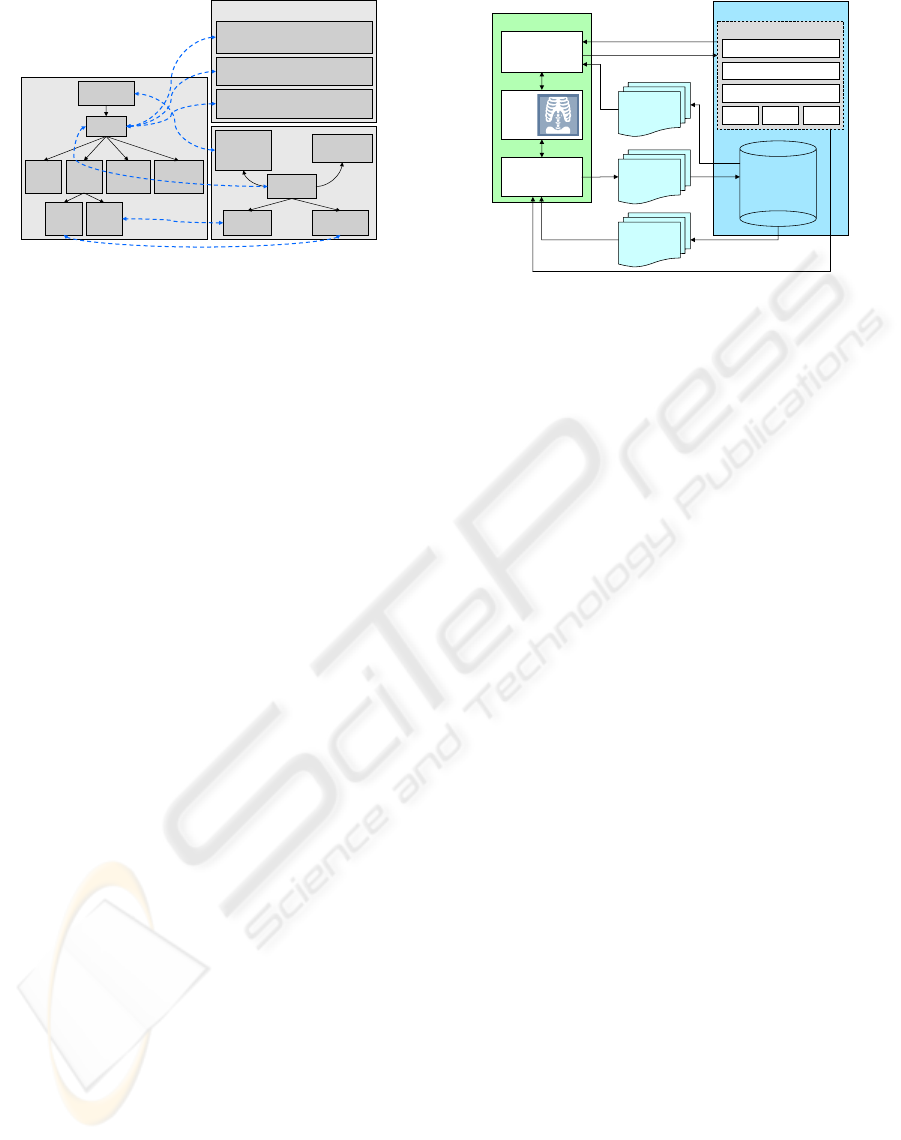
RID1384
Mediastinum
RID:1385
Heart
RID1386
Cardiac
Chamber
RID1393
Heart
Valve
RID1407
Pericardium
RID1398
Myocardium
RadLex
RID1395
Mitral
valve
RID1397
Tricuspid
valve
FMAID:7088
Heart
FMAID:7234
Tricuspid valve
FMAID:7235
Mitral
valve
has part
FMAID:85013
Middle
mediastinal
space
contained in
FMAID:9868
Pericardial sac
attaches to
FMA
ICD9:414.06
Coronary atherosclerosis of native
coronary artery of transplanted heart
ICD9:426.0
Atrioventricular
block complete
ICD9:747.9
Unspecified anomaly of
circulatory system
ICD9
Figure 2: Example of some of the relations between the
ontologies.
as well as between “Heart” and “Mitral Valve”, but
these connections are not further qualified. Qualified
information can be obtained via the mapping to
the FMA. Here we find the special connection
attaches_to from “Heart” to “Pericardial sac”.
Meanwhile the relationship between “Heart” and
“Mitral Valve” is of a different type: “Mitral Valve”
is a part of the “Heart”. The big box on the right side
shows a subset of diseases in ICD9 which are related
to the RadLex concept “Heart”. Once an image is
annotated with the anatomical concept “Heart”, we
can use these connections to constrain the set of
concepts offered as disease annotations.
Semantic Search and Ranking. For the retrieval we
implemented a simple yet efficient ranking algorithm.
If a user searches for images annotated with, for
instance, the concept heart then all images annotated
with exactly this concept are returned as well as
images annotated with concepts which have heart as
an ancestor in the RadLex hierarchy. The depth of the
hierarchy traversed can be set as an user parameter.
The images are ranked in terms of the distance of the
annotated concepts from the original query concept.
Constrain Annotation Dimensions. Initially, the
number of possible annotations for an image is huge.
We are using and 8,389 RadLex and 8,686 different
ICD9 terms. To reduce these numbers, we leverage
the fact, that certain diseases can only occur in certain
organs or body parts. Myocarditis can only occur to
the heart. Our mapping between RadLex and ICD9
provides us with such information.
4 SYSTEM AND EVALUATION
System. Fig. 3 shows the overall architecture of
our system. The annotation workflow consists of
fetching DICOM images from the database, using se-
Annotation
Retrieval
PACS
Images
query concepts
SPARQL query engine
FMA RadLexICD9
Annotation Index
Mappings
Semantics
Semantic PACS
Image Browser
Images +
Semantic
Annotations
result list
domain semantics
Images +
Semantic
Annotations
2D/3D
Image
Viewer
Figure 3: System Architecture.
mantics to annotate the images which are then saved
into corresponding DICOM headers, and finally sav-
ing the semantically enriched DICOM images back
into the database. The retrieval workflow consists of
querying the semantic engine with concepts, which
returns a list of pointers to images clicking any of
which fetches the actual image. The DICOM headers
of the images are parsed to recover the annotations.
Our prototype is implemented in Java using Jena
(http://jena.sourceforge.net) for managing ontologies
and semantic metadata. All ontologies and mappings
were either available in or converted to OWL format.
For the FMA we used the OWL translation by (Noy
and Rubin., 2007). Ontologies and mappings add up
to more than 500 MB data in the triple store. Still, the
combination of Jena and MySQL is providing us with
reasonable response times below two seconds even
for queries with constrains both on the anatomical as
well as on the disease annotations.
To ease the task of finding appropriate annota-
tions we use auto-completing combo-boxes. While
typing in a search term, concept names with match-
ing prefixes are shown in a drop down box and can
be selected. Furthermore, we have also integrated
the Prefuse visualization toolkit (http://prefuse.org)
which displays the neighborhood of concepts and can
be used to easily browse the ontology to identify the
right concepts for annotation. Fig. 1(a) shows the an-
notation front end of the system. The list of disease
terms available in the disease annotation panel can
be constrained by the existing anatomy annotations.
This is done using the mappings between ICD9 and
RadLex.
Fig. 1(b) shows the retrieval front end. The user
can search for anatomical and/or disease concepts
terms. The anatomical terms are based on the RadLex
and FMA vocabulary while the disease terms are
based on ICD9. The user defined depth parameter
limits the extent of the retrieval in the hierarchies
of the ontologies starting from the concept with an
CONTEXT-DRIVEN ONTOLOGICAL ANNOTATIONS IN DICOM IMAGES - Towards Semantic PACS
297
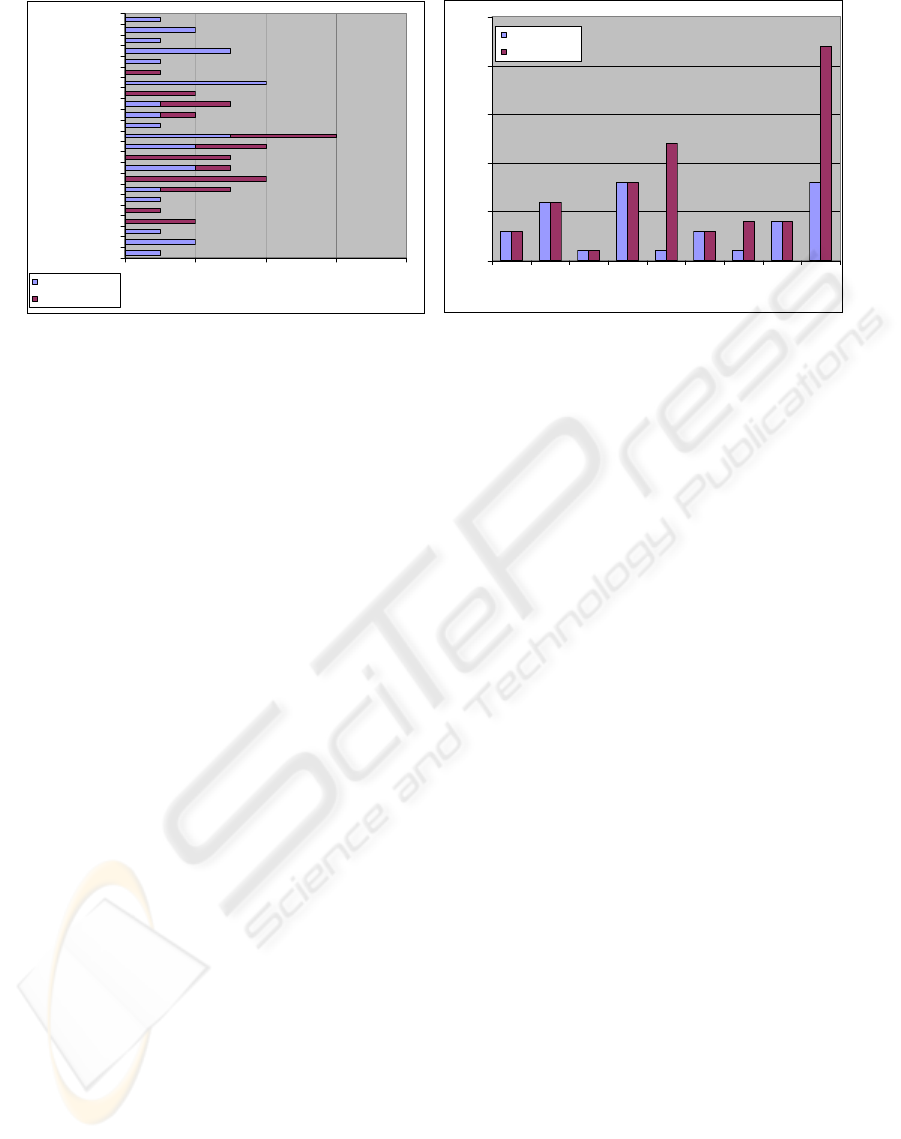
02468
141
375
454
985
1250
1438
1766
2375
processin
g
time (ms)
# terms
# anatomical terms
# disease terms
(a)
0
5
10
15
20
25
123456789
query
# results
semantic depth 0
semantic depth 3
(b)
Figure 4: Evaluation: (a) query processing time vs. number of search terms; (b) search results with/without semantic search.
exact match. For instance, for the query anatomical
concept “heart” and a depth of 3, the search examines
all RadLex concepts which are at a distance up to
3 links away from “heart”. The results are ranked
in sorted order of the distance. Each result consists
of the pointer to the image, the annotations, and the
distance.
Evaluation. We performed preliminary experiments
to evaluate the effectiveness of using semantic con-
cepts in medical images during search. We manually
annotated a set of 45 2D DICOM images with varying
numbers of concepts from both RadLex and ICD9 and
using up to 4 different concepts from each ontology.
Furthermore, we created a set of 23 different queries
each using a different combination of semantic con-
cepts from RadLex and ICD9.
Fig. 4(a) shows the relation between the number
of search terms and the query processing time. The
time measured includes the time for semantic reason-
ing along the RadLex hierarchy, matching with an-
notated concepts, and ranking the results. It was mea-
sured on a 1.8GHz machine with 1GB RAM. Observe
from Fig. 4(a) that certain queries with just anatom-
ical concepts take longer time because of the over-
head of semantic reasoning along the RadLex hier-
archy. Here we perform query expansion which re-
trieves also all images which are annotated with a
subclass of the query concept. The more children
the query concept has, the higher is the effort for the
query expansion. Overall the retrieval is always be-
low 2.5 seconds.
Fig. 4(b) compares search results with and with-
out semantic reasoning along the RadLex hierar-
chy. Clearly, semantic reasoning is useful for certain
queries since it is able to consider a larger space of
potentially relevant images.
We also did a preliminary evaluation on our con-
straining technique using the current RadLex-ICD9
mapping. Constraining was able to limit the space of
possible ICD9 terms for certain RadLex concepts, for
instance, limiting heart to 77, myocardium to 14, and
malleus to 3 ICD9 concepts. Since the technique de-
pends on the RadLex ICD9 mapping, future improve-
ments in the mapping would significantly enrich it.
A demonstration of our prototype to radiologists
at the University Hospital of Erlangen gave us valu-
able feedback. The radiologists liked the idea of split-
ting up the annotation into separate dimensions and
using related context. They also suggested they would
rather have the ability to specify complex queries,
and would even be tolerant of retrieval delays, in-
stead of being limited to simple queries which can be
answered quickly. But in this case the search depth
should be adjustable and the search progress should
be made visible to allow the user to interactively influ-
ence the retrieval process. This is clearly a difference
from typical Web search settings where users often do
not accept retrieval times beyond a few seconds.
Our Prefuse visualization was welcomed as an in-
terface for the exploration of formal domain knowl-
edge in the system which is – if it exists at all – usually
hidden by the application and hard to discover in its
hierarchical structure by the users of existing clinical
applications.
5 DISCUSSIONS
We described techniques for associating formal se-
mantics to medical images and querying and retriev-
ing relevant images using such semantics. Our tech-
niques are based on manually annotating images us-
HEALTHINF 2009 - International Conference on Health Informatics
298

ing semantic concepts from RadLex, FMA and ICD9.
We leverage on developments and standards in the
Semantic Web In contrast to most image annota-
tion work, we investigated serializing the RDF an-
notations to DICOM headers so that they can be di-
rectly archived with their respective images inside to-
day’s PAC systems. Another important contribution
of our work is on making the annotation task sim-
pler and quicker for physicians and radiologists who
typically would be able to devote only minimal time
for such activities. Towards this end, we have created
mappings between anatomical and disease ontologies
such that given annotations from one ontology we can
automatically define the context in the other ontol-
ogy and suggest focused and relevant concepts from
it to the physician for further annotation. Our prelimi-
nary experimental evaluation validates the use of such
context-driven ontological annotation.
The future directions of our work include more
extensive evaluation of the current prototype as well
as exploring possibilities for incorporating better and
different kinds of semantics into the system.
Our current mappings between RadLex and ICD9
as well as between RadLex and FMA are not able
to relate a sizable fraction of concepts between these
ontologies. This is primarily because these concepts
cannot be related by lexical term matching. We are
working on improving the mapping by using fuzzy
string matching techniques, the local graph structures
of the terms in their respective ontologies.
While the system is able to store annotations di-
rectly within DICOM headers, it is still not inte-
grated in an operational PACS environment. This in-
tegration would require modifications to the DICOM
Query/Retrieve service which is the main module re-
sponsible for retrieving images from PACS. We are
working on this integration so that the system can be
easily deployed within current PACS.
Having rich semantic annotations on images
opens up several new dimensions for better medi-
cal image queries. Adding Diagnosis Related Group
codes (http://www.cms.hhs.gov/) will provide a more
holistic semantic view of medical images, since they
cover the classification of different kinds of treat-
ments used by insurance companies. Images can be
subsequently be queried, as an example, for disease
progressions over time. Another application could
be incorporating knowledge from other dimensions,
such as geometric models of organs, for more sophis-
ticated reasoning. We plan to investigate some of
these promising directions as part of a next genera-
tion Semantic PACS platform.
ACKNOWLEDGEMENTS
We would like to acknowledge Sascha Seifert for his
help in DICOM and and the 3D image browser tool.
This research has been supported in part by the THE-
SEUS Program in the MEDICO Project, which is
funded by the German Federal Ministry of Economics
and Technology under the grant number 01MQ07016.
REFERENCES
Buitelaar, P., Sintek, M., and Kiesel, M. (2006). A lexi-
con model for multilingual/multimedia ontologies. In
Proceedings of the 3rd European Semantic Web Con-
ference (ESWC06), Budva, Montenegro.
Carneiro, G., Chan, A. B., Moreno, P. J., and Vasconcelos,
N. (2007). Supervised learning of semantic classes
for image annotation and retrieval. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
29(3):394–410.
Hayes, P. (2004). RDF semantics. W3C Recommendation.
Kahn, C. E., Channin, D. S., and Rubin, D. L. (2006).
An ontology for pacs integration. J. Digital Imaging,
19(4):316–327.
McGuinness, D. L. and van Harmelen, F. (2004). OWL
Web Ontology Language overview. W3C recommen-
dation, World Wide Web Consortium.
Möller, M., Tuot, C., and Sintek, M. (2008). A scien-
tific workflow platform for generic and scalable ob-
ject recognition on medical images. In Tolxdorff,
T., Braun, J., Deserno, T., Handels, H., Horsch,
A., and Meinzer, H.-P., editors, Bildverarbeitung für
die Medizin. Algorithmen, Systeme, Anwendungen,
Berlin, Germany. Springer.
Noy, N. F. and Rubin., D. L. (2007). Translating the Foun-
dational Model of Anatomy into OWL. In Stanford
Medical Informatics Technical Report.
Rosse, C. and Mejino, R. L. V. (2003). A reference on-
tology for bioinformatics: The foundational model of
anatomy. In Journal of Biomedical Informatics, vol-
ume 36, pages 478–500.
Rubin, D. (2007). Creating and curating a terminology for
radiology: Ontology modeling and analysis. Journal
of Digital Imaging, 12(4):920–927.
Rubin, D. L., Mongkolwat, P., Kleper, V., Supekar, K., and
Channin, D. S. (2008). Medical imaging on the se-
mantic web: Annotation and image markup. In AAAI
Spring Symposium Series. Stanford University.
Su, L., Sharp, B., and Chibelushi, C. (2002). Knowledge-
based image understanding: A rule-based production
system for X-ray segmentation. In Proceedings of
Fourth International Conference on Enterprise Infor-
mation System, volume 1, pages 530–533, Ciudad
Real, Spain.
CONTEXT-DRIVEN ONTOLOGICAL ANNOTATIONS IN DICOM IMAGES - Towards Semantic PACS
299
