
WAVELET BASED EXTRACTION OF BLOOD VESSELS
Hammad Omer and Ali Hojjat
Medical Image Computing, KIMHS, University of Kent, Canterbury, CT27PD, U.K.
Keywords: Vessel segmentation, Wavelet coefficients, Image reconstruction, Image enhancement.
Abstract: An algorithm for the segmentation of blood vessels based on the correlation of different wavelet scales is
presented. First the wavelet coefficients are computed for a defined number of scales and then the
correlation between the corresponding coefficients of two consecutive scales is computed. The normalized
product is used as a reference threshold for retaining original wavelet coefficients. If the normalized product
is greater than the corresponding original wavelet coefficient, the original coefficient is retained for image
reconstruction by inverse wavelet transform, otherwise the coefficient is changed with zero value. Low
frequency wavelet coefficients matrix is not used in image reconstruction process as we want only the edge
information. The proposed algorithm is quite general and can be used for the extraction of any type of blood
vessels and provides very promising results.
1 INTRODUCTION
Blood vessel identification and extraction in medical
images is an important step in many medical image
analysis applications e.g. diagnosis of the vessel
stenosis, development of models to analyze different
medical conditions, multimodal image registration
etc. Many vessel extraction techniques have been
proposed in the past. Cemil and Francis (Cemil and
Francis, 2004) presented a very good review of
many such techniques developed in the recent past.
Some of the techniques are suitable for a particular
type of blood vessel extraction e.g. retinal blood
vessels, abdominal blood vessels etc. This limits the
use of these approaches to a particular type of
application only. The vessel segmentation
algorithms developed so far may be broadly
categorized into six main categories (Cemil and
Francis, 2004): 1) pattern recognition techniques, 2)
model-based approaches, 3) tracking based
approaches, 4) artificial intelligence based
approaches 5) neural network based approaches, 6)
tube-like object detection approaches. More details
of these approaches can be found in (Cemil and
Francis, 2004). The blood vessel segmentation
approach presented here is based on correlation of
wavelet coefficients and is based on the idea
presented by Xu (Xu et al., 1994). The approach is
quite general and can be applied to any type of blood
vessels quite confidently.
2 METHODOLOGY
Wavelets constitute a tool to decompose, analyze
and synthesize functions with an emphasis on time-
frequency localization (Omer et al.). Wavelets are
families of functions generated from a single base
wavelet by dilations and translations. The wavelet
coefficient at scale j and time k is calculated as:
∫
+∞
∞−
−= duukuekjWe
j
)()(),(
ψ
(Eq.1)
where
j
ψ
is the wavelet at scale j.
The wavelet transform W(s,t) gives us a scale-
space decomposition of signals and with simple
modifications, images. They help in breaking
complicated signals into simpler components and
can be used in the analysis of complex signals, in the
segmentation or detection of particular features, and
in compression as well as de-noising images. Infact,
wavelets decompose a signal into different
resolution scales.
In a one-level Fast Wavelet Transform (FWT), a
signal C
i
is split into an approximation part C
i+1
and
a detail part D
i+1.
In a multilevel FWT, each
subsequent C
i
is split into an approximation C
i+1
and
detail Di+1. For 2-D images, each C
i
is split into an
approximation C i+1 and three detail channels D
1
i+1,
D
2
i+1,
D
3
i+1
for horizontally, vertically and
diagonally oriented details of the image,
529
Omer H. and Hojjat A. (2009).
WAVELET BASED EXTRACTION OF BLOOD VESSELS .
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing, pages 529-534
DOI: 10.5220/0001558005290534
Copyright
c
SciTePress
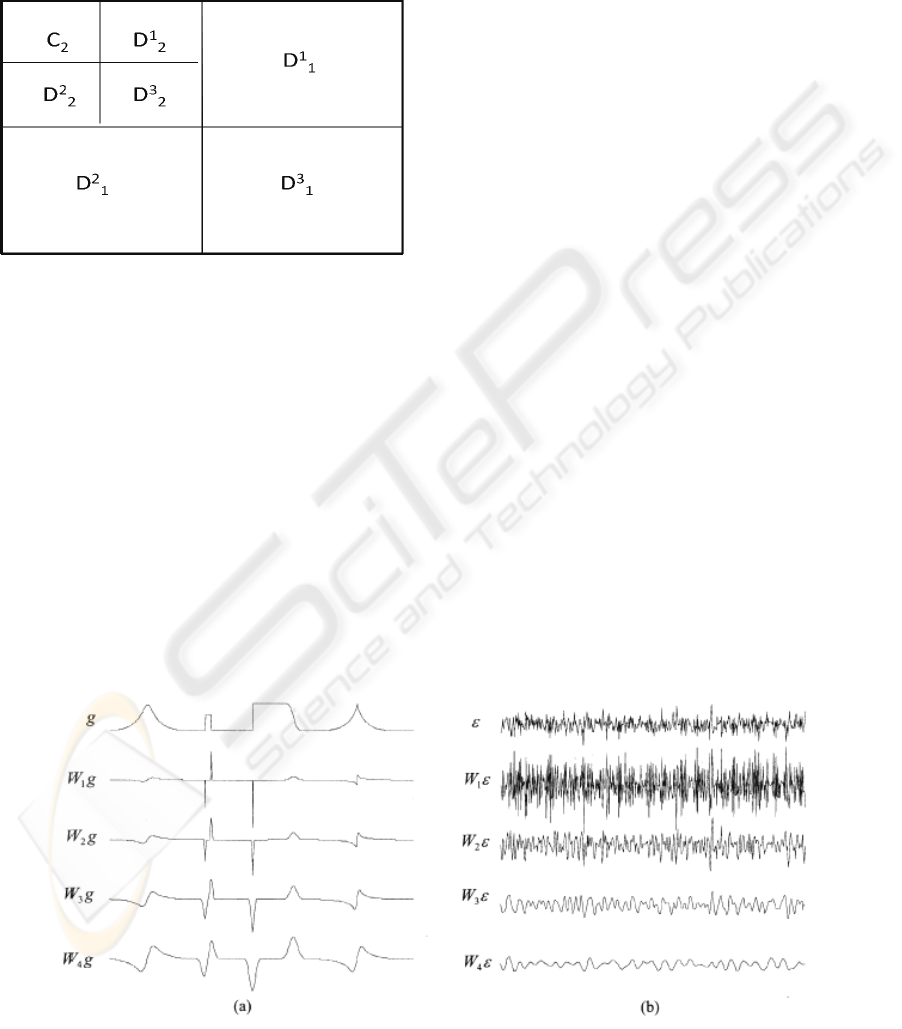
respectively. Figure 1 is an illustration of this
process. The inverse FWT (IFWT) reconstructs each
C
i
from C
i+1
and D
i+1
. This transform and its
inverse are called the Fast Wavelet Decomposition
(FWD) and Fast Wavelet Reconstruction (FWR),
respectively, see (Westenberg and Roderdink, 2000)
for more details.
Figure 1: Ordering of the approximation and detail
coefficients of a two-level 2-D non standard FWT.
Signals and noise behave very differently in
wavelet transform domain. Singularities are more
regular than noise(Xu et al., 1994). The evolution of
singularities and noise across wavelet scales were
analyzed by Mallat et al (Mallat and Hwang, 1992),
(Mallat and Zhong, 1992) and reiterated by Paul Bao
et al (Bao and Zhang, 2003).
As shown in the Figure 2, the edges in a signal
(or image) are represented by large wavelet
coefficients at the corresponding spatial locations
and tend to propagate through the scales. Using a
simple low pass filter would introduce heavy
blurring due to the cutoff of useful components at
the finer scales (at higher frequencies). To retain the
useful high frequency image features as well, it is
very important to distinguish between the high
frequency contributions from the actual signal and
those from the noise. Infact, most of the signal
features contributing to high frequencies also
contribute to low frequencies at the same spatial
locations. Hence there will be correlation between
wavelet coefficients of the useful image signal (such
as edges and spikes) at different scales. In the case
of noise, the correlation is much smaller. For this
reason, the correlation across scales is used to
distinguish between noise contribution and signal
features at high frequencies.
So, wavelets are used for subband decomposition
of a signal (or image). The approach has been to
detect edges directly on the wavelet transform data
algorithm, such as those introduced in (Witkin,
1983), (Fu et al., 2008). Xu(Xu et al., 1994) adopted
the direct multiplication of wavelet transform data
(sub-band decompositions of an image) at adjacent
scales to distinguish important edges from noise and
accomplish the task of removing noise from signals.
In practice, it is sufficient to implement the
multiplication at two adjacent scales. So, the DWT
scale products can be calculated as:
P
j
f(x)=W
j
f(x).W
j
+1
f(x) (Eq.2)
Similarly, for 2D images, the multiscale products
have two components:
P
x
j
f(x,y)=W
x
j
f(x,y) W
x
j
+1
f(x,y) (Eq.3)
P
y
j
f(x,y)=W
y
j
f(x,y) . W
y
j
+1
f(x,y) (Eq.4)
Figure 3 shows the DWT and multi-scale products
of a noisy test signal f where
݂
ൌ݃߳
(Eq.5)
Figure 2: a) DWT of a test signal g at the first four scales. b) The DWT of a sequence of Gaussian white noise at the first
four scales (Bao and Zhang, 2003).
BIOSIGNALS 2009 - International Conference on Bio-inspired Systems and Signal Processing
530

Figure 3: The DWT and multi-scale products of a noisy test signal at the first three scales (Bao and Zhang, 2003).
Although the wavelet transform coefficients of
the original signal ‘g’ are immersed into noise ‘ε’ at
fine scales, they are enhanced in the scale products
P
j
f. The significant features of ‘g’ are more
distinguishable in P
j
f than in W
j
f (Bao and Zhang,
2003).
We calculate the correlation of the wavelet
coefficient across consecutive scales starting from
the first scale to the last. Since there is no down
sampling, the j
th
scale has the same number of
coefficients as the first scale. We also estimate the
noise power at each level by observing the wavelet
transform of a small region of interest corresponding
to background signal. We assume the background
signal contains the same noise components as the
image. The position at which Corr
2
is smaller or
equal to that relative to the estimated noise (taken as
threshold value), the coefficient is changed to zero.
Thus the filtering process consists of, first,
calculating the wavelet decomposition and
correlation between different levels and then, if the
correlation value is lower than a threshold value, the
wavelet coefficient to which it refers is assigned a
value of zero, otherwise it is left unchanged (Bao
and Zhang, 2003). The technique can be considered
as a spatially dependant filter (it can be
demonstrated as a spatially dependant mask); it
spatially selects which part of the data is to be kept
(the edges) and which part of the data to eliminate
(noise); the signal is passed where the wavelet
transform is highly correlated across scales and
suppressed elsewhere. (Xu et al., 1994)
The absence of edges or other significant
features in a localized region of the signal allows the
noisy background to be removed. The thresholding
in wavelet domain over several scales sharpens the
image which results in enhancement of major edges
while suppressing noise. This also improves the
accuracy of locating important edges in images. This
method is simple and performs well on MRA images
of the head.
3 ALGORITHM
The proposed algorithm is based on the
identification of important vessel edges by
correlation between different scales of Wavelets.
The flow chart of the algorithm is shown in Figure 4
and can be discussed in the following steps:
1) The size of the input image is computed.
WAVELET BASED EXTRACTION OF BLOOD VESSELS
531
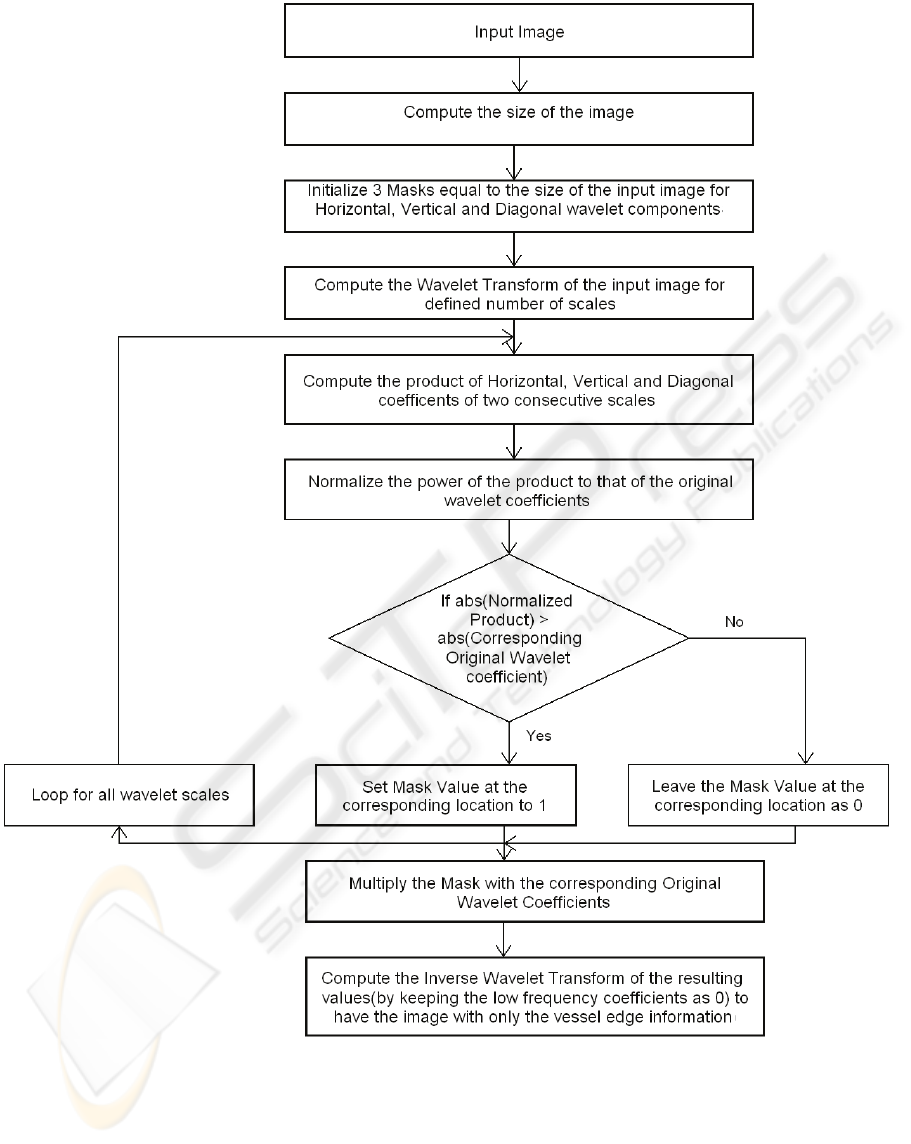
Figure 4: Flow-chart for proposed algorithm for Wavelet Based Vessel Edge Extraction.
2) Three masks are initialized with zero
values. The size of the masks is equal to
that of the original image.
3) The wavelet transform of the input image is
computed for a defined number of scales.
The output of this stage is a set of four
matrices (Low Frequency coefficients, High
Frequency Coefficients in horizontal
direction, High Frequency coefficients in
vertical direction and High Frequency
coefficients in diagonal direction) for each
scale.
BIOSIGNALS 2009 - International Conference on Bio-inspired Systems and Signal Processing
532

Figure 5: [Left] Time of Flight (TOF) MRA of the circle of Willis [Right] Edges enhanced image by the proposed
algorithm.
4) These wavelet coefficients are saved for
later retrieval.
5) The product of the two horizontal detail
coefficient matrices in two consecutive
scales is computed and the same process is
repeated for the vertical as well as diagonal
detail coefficients and for all resolution
levels/scales.
6) The product (i.e. Energy) computed in the
previous step is normalized to that of the
original wavelet coefficients. It enables us
to compare the power of the product to that
of the original wavelet coefficients.
7) A comparison is made between the
normalized product and the original
wavelet coefficients. If the Normalized
product is greater than the corresponding
original wavelet coefficient, the mask value
at the corresponding location is set to 1
otherwise it remains as zero.
8) The masks are multiplied with the original
wavelet and thus only the coefficients
related to edges are retained and others are
lost.
9) The inverse wavelet transform is computed
based on the processed wavelet
coefficients. At this stage, we use only the
processed detail coefficients (i.e.
Horizontal, Vertical and Diagonal) and do
not use the low frequency related wavelet
coefficients.
4 RESULTS AND DISCUSSION
The above algorithm was applied on MRA images
of Circle of Willis and the results obtained were very
good with improved boundaries about blood vessel
edges. The output image showed a clear suppression
of the noisy parts of the image. One important
observation was regarding the number of scales to
be used to find inter-scale correlation which is a
primary measure to identify which wavelet
coefficients belong to the actual signal and which
are representing noise. The use of more than two
scales, causes the loss of much of the edge related
information along with the noisy coefficients and the
reconstructed image does not give very clear
representation of the actual vessel edges. The best
results are obtained with two scales used as almost
all the edges are retained and are very clearly visible
in the resulting image.
Another advantage of this approach is that the
coefficients which are more likely related to noise
are removed during this process of correlation and in
this way, the result is containing noise free edge
information. Figure 5 presents the results of this
algorithm on an MRA image. The left hand side
shows the original MRA image and the image on the
right shows the enhanced image.
5 CONCLUSIONS
We propose a vessel extraction technique based on
normalized inter-scale energy in wavelet domain
which proves to be a very good tool to identify
vessel edges. The noise level in the image is also
WAVELET BASED EXTRACTION OF BLOOD VESSELS
533

reduced. The wavelet coefficients with small
information at the higher scales are removed as they
are more probably associated with noise. The image
reconstruction involves the computation of inverse
wavelet transformation of the processed detailed
coefficients and suppressed low frequency
coefficients. The resulting image contains only the
contours of the blood vessels. This algorithm makes
no assumption about the vessel shapes so it can be
applied to the vessels of any part of the body. The
future work may involve the optimization of the
proposed algorithm. This algorithm is applied onto
MIP (Maximum Intensity Projection) image of Time
of Flight MRI image in which 3D data is mapped on
a 2-D plane. During this process of image
projection, some important information is lost. If this
algorithm could be extended to 3D data, so that the
algorithm would not be applied on 2D projection
image but directly on the 3D image. It will improve
the quality of the reconstructed image. Furthermore,
since 3D involves huge quantity of data to be
processed, introduction of some parallel approach
may considerably reduce the computational time.
REFERENCES
M. A.Westenberg and J. B. T. M. Roderdink, “Frequency
domain volume rendering by the wavelet X-ray
transform,” IEEE Trans. Image Processing, vol.9,
pp.1249-1261, July 2000
S. Mallat and W. L. Hwang, “Singularity detection and
processing with wavelets”, IEEE Trans .Inform.
Theory, vol 3, pp.617-643, Mar.1992
S. Mallat and S. Zhong, “Characterization of signals from
multiscale edges”, IEEE Trans.Pattern Anal. Machine
Intell., vol.14.pp.710-732, July 1992
Paul Bao and Lei Zhang ,”Noise Reduction for Magnetic
Resonance Images via Adaptive Multiscale Products
Thresholding”, IEEE Trans. On Medical Imaging,
vol.22, No.9, September 2003
A. Witkin, “Scale space filtering”, in Proc. 8
th
Int.Joint
Conf. Artificial Intell., 1983
Xu Y., Weaver J. B., Healy D. M. and Lu J. Wavelet
Transform Domain Filters: A spatially selective noise
filtration technique IEEE Trans.Image Process.3 pp.
747-757, 1994
Cemil, Francis, “A Review of Vessel Extraction
Techniques and Algorithms”, ACM Computing
Surveys, Vol.36, No.2, June 2004, pp.81-121
Guoyi Fu, Ali Hojjat and Alan Colchester, “Wavelet Noise
Reduction Based on Energy Features”, ICIAR 2008,
pp.75-84
Hammad Omer, Delakis Ioannis, Kitney Richard,
“Wavelet-based de-noising algorithm for images
acquired with parallel magnetic resonance imaging
(MRI)", Physics in Medicine and Biology, Vol.52,
pp.3741-3751
BIOSIGNALS 2009 - International Conference on Bio-inspired Systems and Signal Processing
534
