FROM INDIVIDUAL INTENSITY VOXEL DATA TO
INTER-INDIVIDUAL PROBABILISTIC ATLASES OF BIOLOGICAL
OBJECTS BY AN INTERLEAVED
REGISTRATION-SEGMENTATION APPROACH
Felix Bollenbeck
1
, Diana Weier
2
, Wolfram Schoor
1
and Udo Seiffert
1
1
Fraunhofer Institute for Factory Operation and Automation (IFF), Sandtorstr. 22, D-39106 Magdeburg, Germany
2
Leibniz Institute of Plant Genetics and Crop Plant Research, Corrensstr. 3, D-06466 Gatersleben, Germany
Keywords:
Registration, Segmentation, 3-D Averaging Atlases, Plant Models.
Abstract:
In this paper we describe an automated processing of plant serial section data for high-resolution 3-D models
of internal structures. The processing pipeline includes standardization and registration of large image stacks
as well as multiple tissue recognition by a joint registration-segmentation approach. By integrating segmented
data from multiple individuals in a common reference, a statistical three-dimensional description is used to
represent the inherent biodiversity amongst specimen. Inter-individual 3-D models are a novelty in the con-
text of plant microscopy, and along with meaningful visualisation they deliver new insights into growth and
development as well as provide a framework for the integration of functional data.
1 INTRODUCTION
The importance and the value of three-dimensional
computer models of tissues or organs can undoubt-
edly be taken for granted. These models often serve as
anatomical atlases facilitating the integration of het-
erogeneous experimental information, such as func-
tional or gene-expression data, with spatial or even
spatio-temporal reference.
The inherently existing inter-individual diversity
leads to a certain divergence between the model and
an arbitrary natural individual. We are working to-
wards statistically valid models by means of barley
grains, based on a multitude of digital 3-D models
from histological serial section data.
The advantages in resolution of serial-section data for
digital models on a micrometer scale generally come
with high costs in 3-D reconstruction (registration)
and labelling (segmentation), since the object of inter-
est is essentially destroyed for digitization, delivering
several thousands of separate and unlabeled raw im-
ages. Existing works for 3-D model generation from
microscopic serial section imaging employ interac-
tive techniques (Gubatz et al., 2007) as well as au-
tomated processing (Dercksen et al., 2008) utilizing
Figure 1: Perspectives of a digital barley grain. A stack of
2,217 individual section images (approx. 4 GB) which was
standardized and registered to recompose an intact grain ob-
ject visualized in a volume rendering, displaying the recon-
structed histology in virtual lateral section.
supervised classification schemes for tissue labelling.
These approaches have in common that models are
generated from data from a single individual, ignor-
ing inter-individual variances in histology and mor-
phology, whereby validity and predictive power for
125
Bollenbeck F., Weier D., Schoor W. and Seiffert U. (2009).
FROM INDIVIDUAL INTENSITY VOXEL DATA TO INTER-INDIVIDUAL PROBABILISTIC ATLASES OF BIOLOGICAL OBJECTS BY AN INTERLEAVED
REGISTRATION-SEGMENTATION APPROACH.
In Proceedings of the Fourth International Conference on Computer Vision Theory and Applications, pages 125-128
DOI: 10.5220/0001767001250128
Copyright
c
SciTePress

new individuals is diminished. Probabilistic models,
reflecting the diversity within biological phenotypes
are highly feasible as reference atlases, apart from the
possibility to quantify phenotypic variance itself.
Since high-throughput processing of section images
is a prerequisite towards sufficiently large data-sets
of anatomical data, we have developed a processing
pipeline for an automated reconstruction of serially
sectioned objects. By utilizing a joint segmentation–
registration algorithm, we address the identification
and labelling of relevant biological structures. In on-
going research we propose an initial modelling based
on labelled voxel-data from a multitude of individuals
for 3-D modelling comprising statistical descriptions
of anatomical diversity and subsequent 3-D visualiza-
tion.
2 INTERLEAVED
REGISTRATION-
SEGMENTATION OF SECTION
DATA
The processing of image data towards inter-individual
models is carried out from individual section images,
to individual image stacks to statistical 3-D diversity
models, where our proposed pipeline is based on in-
terleaved segmentation and registration steps for iter-
ative construction of inter-individual models.
Image Acquisition. For the imaging of functional
units and tissues in microscopic plant organs, speci-
men of barley grains at distinct developmental time
points are prepared for serial sectioning by embed-
ding in a polymer and a contrasting agent. Embed-
ded material is serially sectioned with a microtome
into 3µm thick slices and digitized with a conven-
tional light microscope (1.83 × 1.83µm/px), yielding
roughly 2,000 images per grain, which are stored as
12-bit grayscale image, since contrasting produces lit-
tle color information.
Raw Image Segmentation and Registration. Manual
handling and dust particles on the microscope slides
produce high frequency noise as well as larger dis-
turbances in section images. To remove these dis-
turbances for the subsequent processing, the region-
of-interest (ROI) is segmented, masked, and embed-
ded in a uniform background. We employ a multi-
resolution strategy using a variant of the Level-Sets
approach (Caselles et al., 1997) for active contours
segmentation suggested in (Li et al., 2005).
This initial segmentation of the section object is a
preliminary for using the well-established Principal
Axis Transform (PAT) (Alpert et al., 1990) for uni-
form image moments, since section slices appear in
arbitrary orientation and positioning caused by man-
ual preparation. Employing a PAT thereby serves as
an initialization for the subsequent registration of the
whole image stack (see (Schmitt et al., 2006) for a
comprehensive study), allowing to re-establish three-
dimensional coherence of sectioned objects, which is
lost during sectioning.
Stack Registration and Tissue Recognition. By reg-
istering the full image stack, i.e. finding an optimal
superposition over all images in the stack, the sec-
tioned object is reconstructed. While finding an opti-
mal affine transform, maximizing the correspondence
of all stack images at once using numerical optimiza-
tion schemes is computationally too complex, a pair-
wise sequential alignment of images is error-prone
by propagating possible misalignments through the
stack.
We use a spatially extended intensity-based image-to-
image metric of a w
i
, i = 1,.. .,N weighted sum of
SSD values
1
D(R ◦ ϕ) :=
M
∑
i=2
i−1
∑
j=i−N
w
i− j
Z
Ω
(R
j
(x) ◦ ϕ
j
− R
i
(x) ◦ ϕ)
2
dx
!
= min
(1)
within a local neighborhood N of all M slices (and re-
spective transforms ϕ
j
of images R
j
for positive j) for
more robust stack registration. The whole stack vol-
ume is finally resampled on an isotropic grid.
A uniform alignment of corresponding structures in
the image stack (see fig. 1) in turn allows to re-
late a single section to a reference section using free-
form deformations, which is exploited for the seg-
mentation into prevailing tissues. In the segmenta-
tion step relevant biological structures within an im-
age I : Ω ⊂ R
2
7→ R
+
are recognized and assigned
a unique label S : I ⊂ Ω 7→ {1, .. .,M} for M tissues
or classes. The classification of image grid points
into multiple classes is a crucial step in the modelling
pipeline. Here the raw intensity data is abstracted to-
wards the rationale of the modelling process itself,
where labeled voxel-data is the basis for quantifica-
tion and surface-based modelling of internal struc-
tures. An automatic segmentation of sections is char-
acterized by several requirements:
• A multitude of tissues must be recognized
• Images lack clearly defined edges and structures
1
Here the computational cheap Sum Of Squared Differ-
ences (SSD) is used, because of histogram equalization in
the preprocessing steps.
VISAPP 2009 - International Conference on Computer Vision Theory and Applications
126

• The identification of tissue types needs expert
knowledge
This necessitates the use of algorithms incorpo-
rating a priori information for robust multiclass-
segmentation, where solely intensity-based tech-
niques are clearly unfeasible.
In the context of section imaging in (Bollenbeck
and Seiffert, 2008) we have suggested the segmen-
tation into multiple classes by intensity driven regis-
tration and deformation of reference segmentations,
performing equally accurate with image-feature based
supervised classifiers like support vector machines
and multi layer perceptrons in experiments on histo-
logical plant data, while being less computationally
costly.
Employing the well known free-form deformable reg-
istration formulation
J(u) := D(R,T ; u) + αs(u)
!
= min . (2)
of images R,T : Ω ⊂ R
2
7→ N
+
an a priori reference
segmentation S : R ⊂ Ω 7→ {1,...,M} of R is adapted
to segment T driven by an intensity based image-
metric D subject to a regularized deformation u.
By using this method we classify voxels contained in
the image stack to the respective tissue or material
based on a small set of expert created reference seg-
mentations.
These tissue mappings can be individually visualized
by iso-surface renderings as in (Gubatz et al., 2007)
and (Dercksen et al., 2008), where for valid inter-
individual models multiple individual stacks are the
basis for a statistical three-dimensional description.
Statistical 3-D Models of Barley Grains. Whereas in
the context of histological models, works so far have
neglected biodiversity amongst specimen, the novelty
of our approach is to provide a meaningful description
of diversity amongst multiple specimen in one sin-
gle model, allowing to quantify common themes and
structures for further analysis and to provide a mean-
ingful framework for the integration of functional data
acquired from other individuals.
The quantification of inter-individual variability re-
quires the mapping of data into a common reference
frame allowing the estimation of ubiquitous tissues
and regions of varying tissue composition.
While data sets generally vary in their physical exten-
sion, the task is to capture the composition of internal
structures in terms of estimating a probability to each
spatial coordinate for prevailing tissues based on seg-
mented volume data, rather than constructing averag-
ing surfaces or deformation models.
To obtain a transformation invariant to the actual tis-
sue mapping, data sets are registered into a common
coordinate frame by standardizing first image mo-
ments of the mass-centered intensities of the respec-
tive grayscale volumes of sectioned images (Alpert
et al., 1990; Schmitt et al., 2006).
Instead of using registration approaches maximizing
the correspondence of individual label- or grayscaled
volumes directly with affine mappings, spline, or free-
form deformations, a registration based only on indi-
vidual image statistics can be considered un-biased in
terms of leaving the inter-individual variances unaf-
fected.
For each gridpoint ~x ∈ Ω ⊂ R
3
and tissue M we esti-
mate a probability for ~x belonging to tissue M empir-
ically by
p
~x,M
:=
1
|S|
|S|
∑
i=1
δ(S
i
(~x),M) (3)
from segmented data sets S
i
.
Thereby a closed description of the spatial distribu-
tion of tissues and materials amongst specimen terms
of a mapping P : Ω 7→ R
M
is obtained.
While the spatial distribution of tissue probabilities is
not based on assumptions on underlying distribution
as with statistical deformation models, it can directly
be related to underlying histological information (in-
tensity volumes) as indicated in fig. 2.
3 RESULTS
For this work four individual grains at the same de-
velopmental stage were sectioned and digitized as de-
scribed, yielding 2,128 to 2,736 slice images, each of
size 1200 × 1600 pixels (approx. 30 GB image data).
Providing the basis for further modelling, intact indi-
vidual grains were reconstructed from section images
as proposed. Fig. 1 shows a volume rendering of a
registered individual grain. By virtual lateral sections,
revealing reconstituted histological features, such as
cell walls etc., the performance of the registration ap-
proach in processing large image stacks could be ini-
tially validated, while detailed assessment is feasible,
yet beyond the scope of this paper. Employing the
described registration-segmentation algorithm, inten-
sity stacks were segmented into their respective tis-
sues based on a set of reference data defined by an
expert.
For inter-individual description we registered and
joined the segmented data in a common reference,
yielding a volume of probabilities for each tissue (see
fig. 2(a)). Using the probabilistic modelling, we ad-
dressed the biological questions (1.) how are specific
tissues and relevant materials varying and (2.) what
are ubiquitous themes amongst individuals. Thus, for
an insightful 3-D visualization of probabilistic models
FROM INDIVIDUAL INTENSITY VOXEL DATA TO INTER-INDIVIDUAL PROBABILISTIC ATLASES OF
BIOLOGICAL OBJECTS BY AN INTERLEAVED REGISTRATION-SEGMENTATION APPROACH
127
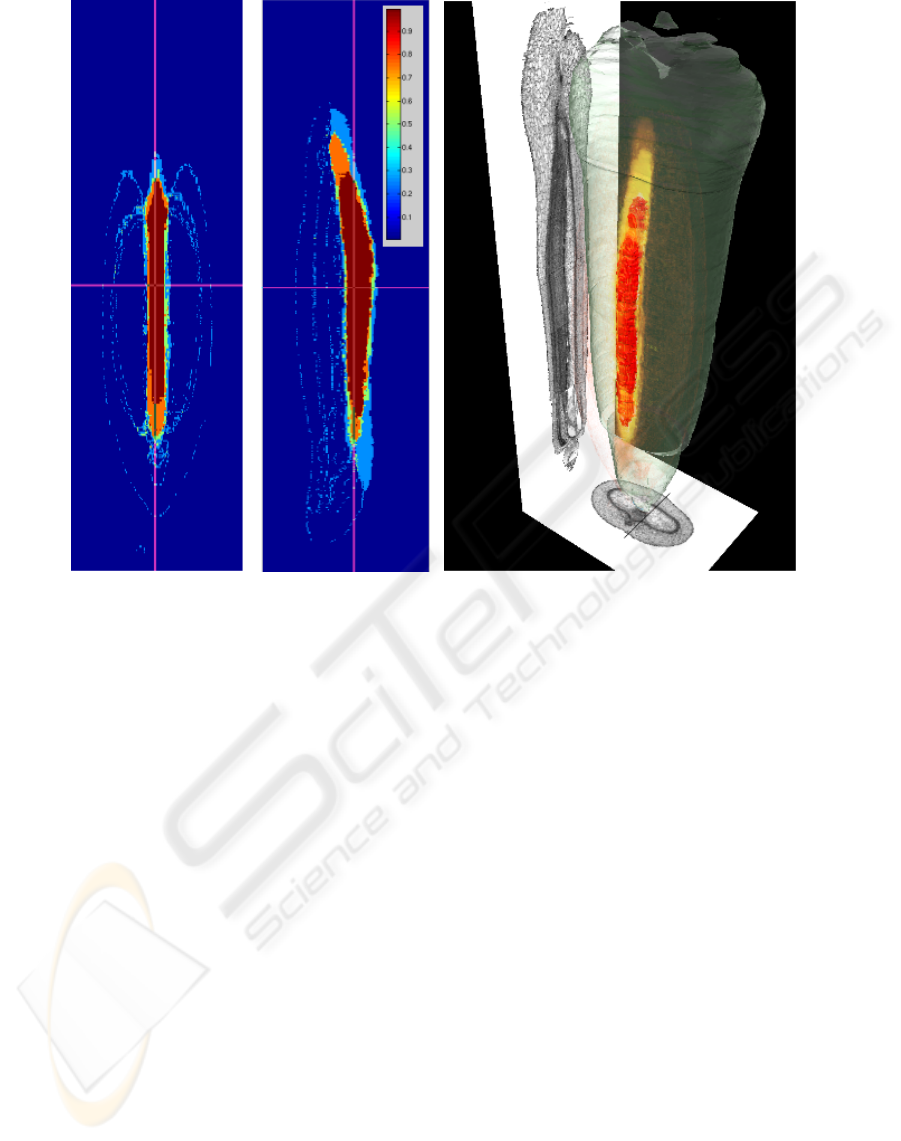
(a) (b)
Figure 2: Visualization of the inter-individual statistical atlas: 2(a) Two orthogonal length-sections through a volume of
position-specific probabilities for the nucellar projection. 2(b) 3-D Rendering of a statistical model for the nucellar projection:
Red volume represents tissue-material ubiquitous to all individuals., the opacity of the yellow volume rendering is proportional
to the probability for the tissue amongst all individuals (projectional view and outer hull from a single individual).
or atlases, we are using a combination of two meth-
ods:
1. Volume rendering for the spatial distribution of
tissue probability values
2. Surface rendering for ubiquitous regions, i.e.
p
~x,M
= 1
Fig. 2 shows a combined rendering for the nucel-
lar projection, which plays an important role in early
grain development. A projectional view of a virtual
lateral and section slice and transparent outer hull of
an individual grain is displayed for better intuition.
Visual analysis revealed that connected regions exist
even for volumetrically small tissues such as the vas-
cular bundle and transfer cells, with small variability
amongst specimen, suggesting a determinant role in
grain development.
4 DISCUSSION AND OUTLOOK
In this paper we present an interleaved registration
and segmentation approach for automated 3-D model
generation by integrating data from multiple indi-
viduals. By this inter-individual processing, high-
resolution statistical models of internal structures are
constructed towards high quality phenotyping of mi-
croscopic plant organs.
This significant extension of existing works on 3-D
histological models provides the basis for system-
atic quantification of phenotypical properties, which
is considered an urging topic in current plant biology.
The presented modelling and quantification of inter-
individual diversity further allows a reliable integra-
tion of functional data into a spatial atlas, whereas the
description of diversity itself might also lead to new
indications of functional interrelationships. Insight-
ful visualization helps to identify common themes
in morphology of developing seeds, especially struc-
tures related to storage compound aggregation.
Currently efforts for statistical models resolved on a
VISAPP 2009 - International Conference on Computer Vision Theory and Applications
128

timeline are underway: For such digital morphogene-
sis population–averaging models are a preliminary.
ACKNOWLEDGEMENTS
This work is supported by BMBF grant 0313821. The
authors want to thank U. Siebert and B. Zeike (IPK
Gatersleben) for the material preparation, R. Pielot,
and W. Weschke (IPK Gatersleben) for fruitful dis-
cussions and useful comments.
REFERENCES
Alpert, N. M., Bradshaw, J. F., Kennedy, D., and Correia,
J. A. (1990). The principal axes transformation - a
method for image registration. The Journal of Nuclear
Medicine, 31:1717–1722.
Bollenbeck, F. and Seiffert, U. (2008). Fast registration-
based automatic segmentation of serial section images
for high-resolution 3-d plant seed modeling. Biomed-
ical Imaging: IEEE 5th Int. Symp., pages 352–355.
Caselles, V., Catte, F., Coll, T., and Dibos, F. (1997). A geo-
metric model for active contours in image processing.
Int. J. Comput. Vision, 22:158–175.
Dercksen, V. J., Br
¨
uß, C., Stalling, D., Gubatz, S., Seiffert,
U., and Hege, H.-C. (2008). Towards automatic gen-
eration of 3D models of biological objects based on
serial sections. In Linsen, L., Hagen, H., and Hamann,
B., editors, Visualization in Medicine and Life Sci-
ences, pages 3–25. Springer.
Gubatz, S., Dercksen, V., Br
¨
uß, C., Weschke, W., and
Wobus, U. (2007). Analysis of barley (Hordeum vul-
gare) grain development using three-dimensional dig-
ital models. The Plant Journal(12), 52(4):779–790.
Li, C., Xu, C., Gui, C., and Fox, M. (2005). Levelset set
evolution without re-initialization: A new variational
formulation. In Proc. of the CVPR.
Schmitt, O., Modersitzki, J., Heldmann, S., Wirtz, S., and
Fischer, B. (2006). Image Registration of Sectioned
Brains. Int. J. Comput. Vision, 73(1):5–39.
FROM INDIVIDUAL INTENSITY VOXEL DATA TO INTER-INDIVIDUAL PROBABILISTIC ATLASES OF
BIOLOGICAL OBJECTS BY AN INTERLEAVED REGISTRATION-SEGMENTATION APPROACH
129
