
AUTOMATIC DATA EXTRACTION IN ODONTOLOGICAL X-RAY
IMAGING
Douglas E. M. de Oliveira
1
, Gilson A. Giraldi
1
, Luiz A. Pereira Neves
2
Adriana G. da Costa
3
and
´
Erika C. Kuchler
3
1
National Laboratory for Scientific Computing, Av. Getulio Vargas, 333, Petr
´
opolis, Brazil
2
State University of Santa Catarina, Vision Laboratory, S
˜
ao Bento do Sul, Brazil
3
Federal University of Rio de Janeiro, RJ, Brazil
Keywords:
Thresholding, Mathematical Morphology, PCA, Feature Extraction.
Abstract:
Automating the process of analysis in dental x-ray images is receiving increased attention. In this process,
teeth segmentation from the radiographic images and feature extraction are essential steps. In this paper, we
propose an approach based on thresholding and mathematical morphology for teeth segmentation. First, a
thresholding technique is applied based on the image intensity histogram. Then, mathematical morphology
operators are used to improve the efficiency of the teeth segmentation. Finally, we perform the boundary
extraction and apply the Principal Component Analysis (PCA) to get the principal axes of the teeth and some
lengths along it that are useful for dentist diagnosis. The technique is promising and can be extended for other
applications in dental x-ray imaging.
1 INTRODUCTION
Automating the procedure of image analysis for x-ray
dental images is an important tool for diagnosis and
planning of dentistry procedures. From the viewpoint
of image processing, two problems are fundamental
in this process: segmentation and feature extraction.
From a practical point of view, segmentation is
the partition of an image into multiple regions (sets
of pixels) according to some criteria of homogeneity
of features such as color, shape, texture and spatial
relationship (Jain, 1989). These fundamental regions
are disjoint sets of pixels and their union compose
the original whole scene. Approaches in image seg-
mentation can be roughly classified in: (a) Contour
Based methods, like snakes and active shape models
(Suri et al., 2002; Kass et al., 1988); (b) Region based
techniques (Suri et al., 2005); (c) Global optimiza-
tion approaches (Pan, 1994); (d) Clustering methods,
like k-means, Fuzzy C-means, Hierarchical clustering
and EM (Zhu and Yuille, 1996); and (e) Thresholding
methods (Albuquerque et al., 2004).
Among these approaches, thresholding techniques
(compute a global threshold to distinguish objects
from their background) are simple for implementa-
tion, with low computational cost, been effective tools
to separate objects from their backgrounds (Sahoo
et al., 1988). These methods have been successfully
applied for document image analysis, scene process-
ing, quality inspection, and medical imaging. The
common approach to implement a thresholding tech-
nique is based on the image histogram by searching
for its local minima (valleys). Other possibility is to
search for a threshold value constrained to the max-
imization of some information measure or entropy,
like the classical Shannon or generalizations of it (Al-
buquerque et al., 2004). After performed the image
binarization through the obtained threshold, we can
apply mathematical morphology techniques in order
to improve the result (Rodrigues et al., 2006).
Once the geometry of the objects has been ex-
tracted we can proceed the feature extraction. For
instance, geometric features are of special interest in
this project. Contour-based features, like area and cir-
cularity, as well as anatomical features can be used for
information extraction and classification (Rodrigues
et al., 2006; Jain and Chen, 2004).
In this paper, we propose an approach for teeth
segmentation and feature extraction which is based on
the following steps. First, a thresholding technique
is applied based on the image intensity histogram.
Then, mathematical morphology techniques are used
141
E. M. de Oliveira D., A. Giraldi G., A. Pereira Neves L., G. da Costa A. and C. Kuchler É. (2009).
AUTOMATIC DATA EXTRACTION IN ODONTOLOGICAL X-RAY IMAGING.
In Proceedings of the First International Conference on Computer Imaging Theory and Applications, pages 141-144
DOI: 10.5220/0001800601410144
Copyright
c
SciTePress
to complete the teeth segmentation. Next, we perform
the feature extraction. The teeth boundary is extracted
from the binarized image and the Principal Compo-
nent Analysis (PCA) (Fukunaga, 1990) is used to get
the principal axes (r) of the teeth. Next, we automat-
ically determine two measures along r: the crown-
body (CB) and root (R) lengths. The former is ob-
tained through the distance from the deepest pit to the
furcation. The latter is the distance from the furca-
tion to the root apex. Finally, we compute the ration
CB/R.
The results show that the technique is promising
and can be extended for other applications in dental
x-ray imaging.
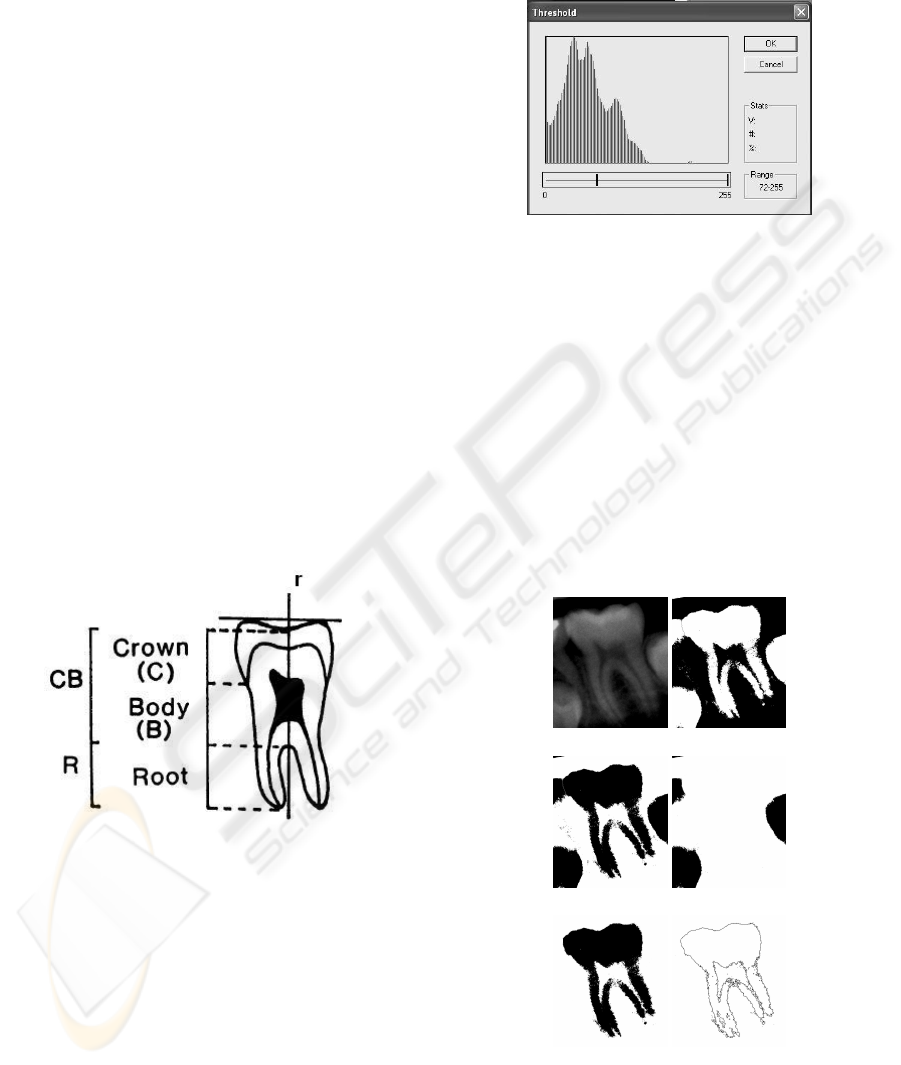
2 PROPOSED METHOD
In this section we describe the main steps of our tech-
nique for segmentation and information extraction in
dental x-ray images. The segmentation step is based
on a thresholding method and mathematical morphol-
ogy operators. Information extraction is performed
by contour detection and PCA. The Figure 1 pictures
a generic teeth boundary and the target features: prin-
cipal axes (r), deepest pit (C), furcation (B) and root
apex (R).
Figure 1: Teeth features: principal axes (r), deepest pit (C),
furcation (B) and root apex (R).
In the section 2.1 we describe the pipeline for
boundary extration and in section 2.2 the information
extraction step is presented.
2.1 Mathematical Morphology and
Boundary Extraction
A typical histogram for the dental images is pic-
tured on Figure 2. We have observed that the second
and third local maxima gives the intensity range that
roughly covers the structure of interest. For instance,
Figure 2 shows the histogram for Figure 3.a and the
Figure 3.b shows the thresholding result.
Figure 2: Typical histogram for the data base images.
We observe some undersegmentation in this fig-
ure. Then, we invert the image and starts the pro-
cess to generate a mask in order to discard wrong pix-
els. A simple search method is used to get the mask
(Figure 3.d). This procedure is done using gray scale
histogram thresholding. Thus, it is possible to iden-
tify the tooth crowns and remove the main tooth. So,
a XOR operation is performed with the inverted im-
age, giving the result pictured on Figure 3.e. Finally,
an opening operation extracts the boundary pixels, as
shown in Figure 3.f. This operation is done using
cross structuring element with a single iteration.
(a) (b)
(c) (d)
(e) (f)
Figure 3: Morphological Chain: (a) Original image. (b)
Thresholding result. (c) Inverted image. (d) Mask. (e) XOR
between Figures 3.c and 3.d. (f) Boundary pixels.
IMAGAPP 2009 - International Conference on Imaging Theory and Applications
142

2.2 Information Extraction
Once performed the boundary extraction, we must
consider geometric features. The main axes of the
teeth is the first target in this step. As already known
in the literature (Fukunaga, 1990), it can be obtained
by the Principal Component Analysis (PCA), also
called Karhunen-Loeve, or KL method (Jain, 1989).
Thus, let us suppose that the data consists of N tuples
or data vectors, from a n-dimensional space. Then,
PCA searches for k n-dimensional orthonormal vec-
tors that can best be used to represent the data, where
k ≤ n, in the sense of data compression. Figure 4 pic-
ture this idea using a bidimensional representation. If
we suppose that the data points S =
{
u
1
,u
2
,...,u
N
}
are distributed over the teeth boundary, it follows that
the coordinate system
X,Y
, shown in Figure 4, is
more suitable for representing the data set. The PCA
technique gives this coordinate system through the
following steps (Fukunaga, 1990):
1. Compute the centroid of the data set:
C
M
=
1
N
N
∑
i=1
u
i
. (1)
2. Subtract the centroid C
M
from the data points:
y
i
= u
i
− C
M
,i = 1, 2, · · ·, N. (2)
3. Compute the covariance matrix R :
R =
N
∑
i=1
y
i
y
∗T
i
. (3)
4. Find the matrix
Φ = [Φ
1
,Φ
2
,· · ·,Φ
n−1
,Φ
n
] (4)
where Φ
i
is the i-th eigenvector of R, sorted in de-
creasing order of the corresponding eigenvalues.
The columns of Φ gives the new basis vectors.
The operation:
z
i
= Φ
T
y
i
, i = 1, 2, ..., n, (5)
computes the data representation in the new basis.
Therefore, we take the obtained curve (boundary), ap-
ply steps (1)-(4) to get Φ, a 2 × 2 matrix in our case
(n = 2). Then, Φ
1
is taken as the principal axes of the
teeth (Figure 4). Expression (5) is applied for chang-
ing coordinates in order to simplify the searching for
the deepest pit, the furcation and the root apex (Figure
1).
Figure 4: (a)Original dataset. (b) Extraction of the principal
component.
3 DISCUSSION
We have a data base composed by 150 panoramic ra-
diographs, with resolution of 300 dpi, in gray scale
that will be used to test the method. The obtained re-
sults will be compared with manual evaluations per-
formed by the radiologists of our team. So, some
statistics are going to be generated in order to quantify
the precision of the proposed method.
Region based and contour based approaches could
be also considered to perform the segmentation tasks.
However, region based methods depends on some
characterization of the intensity patterns inside the re-
gion of interest (Suri et al., 2005; Zhu and Yuille,
1996). It is not a simple task due to inhomogeneities
of the intensity field as well as texture patterns in-
side the image, as we can observe in Figure 3.a. On
the other hand, contour based approaches, like snakes
or level set models (Shah et al., 2006; Keyhaninejad
et al., 2006), must be properly parameterized in or-
der to converge to the desired boundary without stop-
ping on local minima. By considering that the teeth
boundary follows some pattern, a deformable model
incorporating shape information (shape model) could
be more efficient than a traditional (free) snake model
(Buchaillard et al., 2007).
4 CONCLUSIONS
The proposed method is a combination of mathe-
matical concepts in morphology and shape analysis
AUTOMATIC DATA EXTRACTION IN ODONTOLOGICAL X-RAY IMAGING
143

(PCA), as well as algorithms to automatically com-
pute the ratio CB/BR in x-ray images. The computer
implementation has been developed using Object Ori-
ented best practices.
REFERENCES
Albuquerque, M. P., Albuquerque, M. P., Esquef, I., and
Mello, A. (2004). Image thresholding using tsallis en-
tropy. Pattern Recognition Letters, 25:1059–1065.
Buchaillard, S. I., Ong, S. H., Payan, Y., and Foong, K.
(2007). 3d statistical models for tooth surface recon-
struction. Comput. Biol. Med., 37(10):1461–1471.
Fukunaga, K. (1990). Introduction to statistical patterns
recognition. Academic Press, New York., 18(8):831–
836.
Jain, A. K. (1989). Fundamentals of Digital Image Process-
ing. Prentice-Hall, Inc.
Jain, A. K. and Chen, H. (2004). Matching of dental x-ray
images for human identification. Pattern Recognition,
37(7):1519–1532.
Kass, M., Witkin, A., and Terzopoulos, D. (1988). Snakes:
Active contour models. International Journal of Com-
puter Vision, 1(4):321–331.
Keyhaninejad, S., Zoroofi, R. A., Setarehdan, S. K., and
Shirani, G. (2006). Automated segmentation of teeth
in multi-slice ct images. In Visual Information Engi-
neering, pages 339–344.
Pan, H. (1994). Two-level global optimization for image
segmentation. ISPRS journal of photogrammetry and
remote sensing, 49(2):21–32.
Rodrigues, P. S., Giraldi, G. A., Chang, R. F., and Suri,
J. (2006). Non-extensive entropy for cad systems of
breast cancer images. In Society, I. C., editor, Pro-
ceedings of Brazilian Simposium on Computer Graph-
ics and Image Processing, pages 121–128, Belo Hori-
zonte, Brazil.
Sahoo, P. K., Soltani, S., and Wong, A. K. C. (1988). A sur-
vey of thresholding techniques. Comput. Vis. Graph-
ics Image Process, 41:233–260.
Shah, S., Abaza, A., Ross, A., and Ammar, H. (2006). Auto-
matic tooth segmentation using active contour without
edges. In Proc. of the Biom. Consortium Conference,
pages 1–6.
Suri, J. S., Liu, K., Singh, S., Laxminarayan, S., Zeng,
X., and Reden, L. (2002). Shape recovery algorithms
using level sets in 2-d/3-d medical imagery: a state-
of-the-art review. IEEE Transactions on Information
Technology in Biomedicine, 6(1):8–28.
Suri, J. S., Wilson, D., and Laxminarayan, S. (2005). Hand-
book of Biomedical Image Analysis: Volume 3: Reg-
istration Models (International Topics in Biomedical
Engineering). Springer-Verlag New York, Inc.
Zhu, S. C. and Yuille, A. (1996). Region competition:
Unifying snakes, region growing, and bayes/mdl for
multibang image segmentation. IEEE Trans. Pattern
Anal. Mach. Intell., 18(9):884–900.
IMAGAPP 2009 - International Conference on Imaging Theory and Applications
144
