
A TOOL BASED ON WEB SERVICES TO QUERY
BIODIVERSITY INFORMATION
Joana G. Malaverri, Bruno S. C. M. Vilar and Claudia B. Medeiros
Institute of Computing, University of Campinas, 13083-970, Campinas, SP, Brazil
Keywords:
Biodiversity Systems, Ontologies, Query processing, Web services.
Abstract: Biodiversity Information Systems are complex software systems that present data management solutions to
allow researchers to analyze species and their interactions. The complexity of these systems varies with the
data handled, users targeted and environment in which they are executed. An open problem to be faced
especially in a Web environment is data heterogeneity, and the diversity of user vocabularies and needs. This
hampers query processing. This paper presents a tool based on Web services to expand and process biodiversity
queries using ontology information. This solution relies on a new database organization, also described here,
which combines in a single model data collected in the field with data found in archival sources. This tool is
being tested using real case studies, within a large Web-based biodiversity system.
1 INTRODUCTION
Biodiversity studies cover a wide variety of data, in-
cluding species occurrence records, spatial, ecologi-
cal, socio-economic data and others. The large vol-
ume of information on species and their habitats re-
quires new solutions for managing and analyzing the
characteristics of species and their interactions.
Biodiversity Information Systems emerged with
this objective. The scope of these systems includes
the recovery of textual information, such as literal
descriptions, and of the spatial distribution of one
or more species. Typically, they provide support to
queries on traditional database systems, and users are
limited in query flexibility. Moreover, there is a need
for new tools to process biodiversity data on the Web.
This paper discusses our proposal to this problem
– a tool based on a set of Web services that processes
queries, extending them with semantic information.
This proposal is being tested on the BioCORE project,
a Web biodiversity system that is being developed in
a joint effort between computer scientists and bio-
logists. Our queries are centered on two kinds of
biodiversity data: ocurrence records, containg ob-
servations recorded and collected during field trips;
and catalog records, containing information on (pre-
served) species in museums. This combination of data
sources is itself a contribution, since most biodiver-
sity systems consider either one or the other, but not
both. Our solution combines Web services, query ex-
pansion mechanisms based on ontologies, and a novel
biodiversity database model.
2 RELATED WORK
2.1 Managing Biodiversity Information
and Standards
There are a large number of projects that aim to de-
velop mechanisms to publish and manage biodiver-
sity data on the Web. Data heterogeneity is one of
the most important problems considered. Many of
these projects were proposed in order to manage col-
lections of Museums of Natural History and Herba-
riums. SpeciesLink (CRIA, 2001), for example, is a
Web system that allows integration of information on
biodiversity records available in museums, herbaria
and microbiological collections by publishing them
in the Internet. Another example is Specify (Beach,
2007), a project that aims to provide a platform that
uses Web services as a support for the management
of data collections. It also considers operations that
should be performed on the collections, such as loans,
exchanges, and donations.
On the other hand, the Biota project (Colwell,
1996) was one of the first projects interested in occur-
rence records – those that register observations made
by biologists in the field.
In parallel, projects like GBIF (Global Biodiver-
sity Information Facility) (GBIF, 2004), ITIS (Inte-
305
Malaverri J., Vilar B. and Medeiros C.
A TOOL BASED ONWEB SERVICES TO QUERY BIODIVERSITY INFORMATION.
DOI: 10.5220/0001836103050310
In Proceedings of the Fifth International Conference on Web Information Systems and Technologies (WEBIST 2009), page
ISBN: 978-989-8111-81-4
Copyright
c
2009 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved

grated Taxonomic Information System) (ITIS, 2007),
or TDWG (Taxonomic Database Working Group)
(TDWG, 1994), are directing efforts to establish stan-
dards and infrastructure for integration and interope-
rability of data from biological collections, making
them available on the Web. Another considerable set
of biodiversity applications deals with the manage-
ment of taxonomic information and geographic dis-
tribution of species – e.g., Tree of Life (Maddison and
Schulz, 2007).
Most of these projects use data standards to facil-
itate access and dissemination of information on the
Internet. The two standards that are most commonly
adopted are Darwin Core and ABCD (Access Biolog-
ical Colections Data) (TDWG, 1994). The main ob-
jective of Darwin Core is to facilitate the exchange of
information on species. Among its core attributes, it
specifies the name of the organism and where, when
and who collected it. ABCD brings additional ele-
ments to those provided by Darwin Core. It is a co-
mmon data schema that allows to structure and spec-
ify units of biological collections.
Data transfer protocols like DiGIR (Source-
Forge.NET,1999) and BioCase (BioCase, 2005) were
developed for these standards. DiGIR is a protocol
that provides a single access point to distributed data
sources, and uses the Darwin Core standard. Bio-
Case was developed to provide connectivity between
databases of biological collections. This protocol is
based on HTTP and XML and uses the ABCD stan-
dard to transmit data over the BioCase network. A
new approach known as Tapir (TDWG Access Pro-
tocol for Information Retrieval) is being promoted
by GBIF to enhance interoperability among biodiver-
sity tools and data to unify the DiGIR and BioCASE
protocols and to improve the interoperability between
them. Tapir (TDWG, 1994) specifies a standard pro-
tocol that is based on XML schema and Web services.
Several of these efforts are begining to consider on-
tologies as a means to enhance interoperability in Bio-
diversity Systems.
2.2 Ontologies
An ontology is a specification of a conceptualization
(Gruber, 1993). Ontologies can capture the semantics
of a domain by defining concepts and their relation-
ships. Besides this, it is possible to find specific appli-
cations of ontologies such as description of resources
and services to automate processes, to control vocab-
ularies, contextualize and infer information, etc.
Particularly in biodiversity information systems,
it is possible to find different uses for ontologies.
SEEK (Michener et al., 2007) or Aonde (Daltio and
Medeiros, 2008) use ontologies to enable query and
analysis of the data in multiple and heterogeneous in-
formation sources.
2.3 Query Processing Issues
Our work concerns combining the flexibility of Web
services with mechanisms for modification of biodi-
versity queries to enhance their semantics. Different
query modification techniques can be found in the li-
terature, such as reformulation, expansion, substitu-
tion, enrichment and relaxing e.g., (Florescu et al.,
1996; Lian et al., 2007). The goals of these techniques
include:
• Better performance - e.g., less time or fewer re-
sources needed in query execution;
• Better precision in the results through the modifi-
cation of a query that originally does not retrieve
all relevant results.
Query expansion/rewriting – the technique we
adopted – is the process to augment a user query with
additional terms, to improve results. The techniques
and resources used to expand the queries include on-
tologies and probabilistic methods (Andreou, 2005),
and term extraction through a set of documents ob-
tained or query logs. The use of ontologies corre-
sponds to the so-called Semantic Query Optimization,
which reformulates a query into another, in a more ef-
ficient way, which is semantically equivalent, provid-
ing the same answer (Necib and Freytag, 2004).
3 QUERYING BIODIVERSITY
DATA
3.1 The BioCORE Project
BioCORE (Bio-CORE, 2008) is a Web based project
developed in a collaboration between researchers in
Computer Science and Biology. It aims to aid scien-
tists and researchers in biodiversity to perform multi-
modal and exploratory queries among heterogeneous
biodiversity data sources. Its architecture, presented
on Figure 1, is based on Web services.
The architecture covers a client application,
which supplies an interface between the users and the
provided services. Services are categorized as stora-
ge, support and advanced. The first group provides
basic data access facilities, encapsulating data repo-
sitories at the storage level. Supporting services in-
clude: content based image retrieval, and manage-
ment of collection data, metadata, geographic data
WEBIST 2009 - 5th International Conference on Web Information Systems and Technologies
306
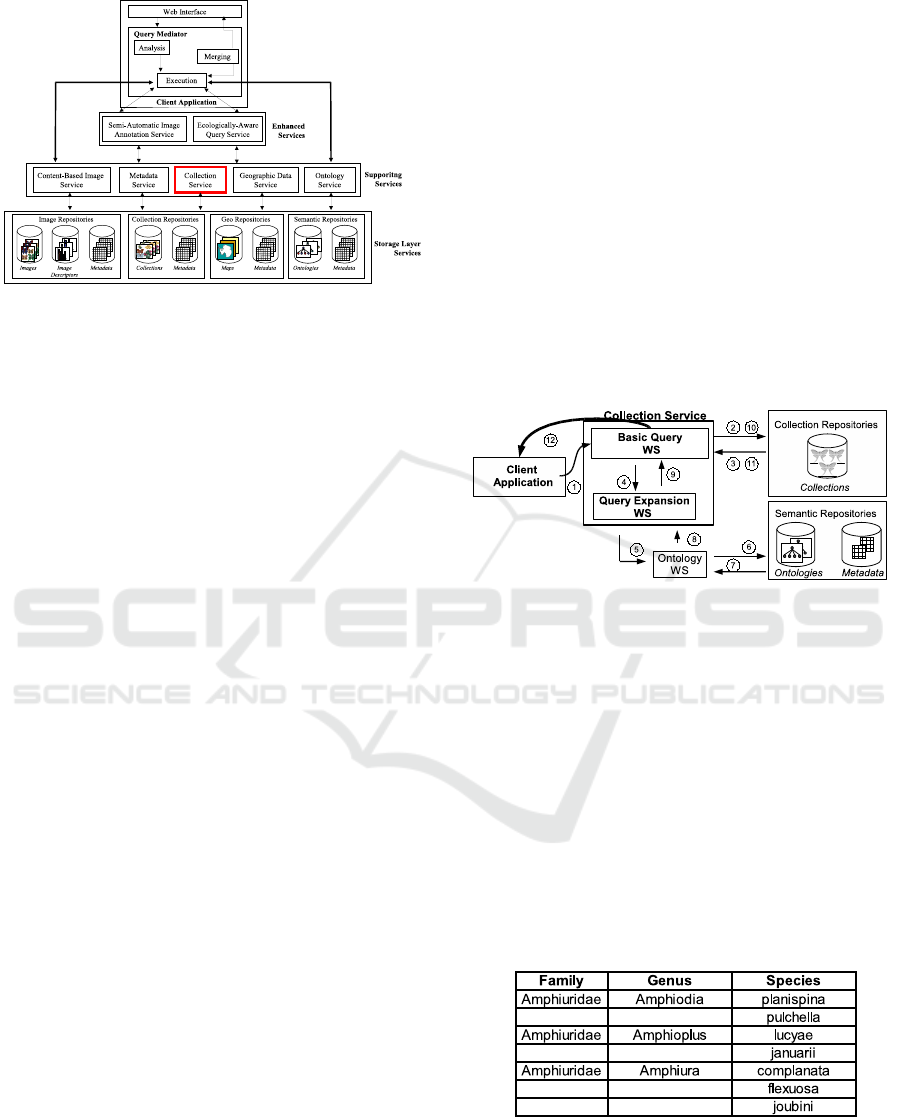
Figure 1: BioCORE Architecture.
and ontologies. Advanced services comprise more
complex services which invoke compositions of sup-
porting services. This paper concerns the Collection
Service, outlined in the figure.
Repositories contain information on images,
maps, collections and ontologies. The latter, stored
in a Semantic Repository, are used by our Collection
service. Also, each repository maintains a set of meta-
data to aid information management and retrieval.
3.2 The Collection Service
The Collection Service is a tool based on Web ser-
vices to query biodiversity records. Its queries are
performed on data stored in Collection Repositories,
and extended by ontologies in Semantic Repositories
(see figure 1).
It is composed of two main elements: (i) a basic
query Web service that receives and executes query
requests from a Client Aplication and (ii) a query ex-
pansion Web service that overwrites a query expres-
sion using ontologies delivered by our Ontology Ser-
vice (Daltio and Medeiros, 2008).
The main features of this tool are: (i) use of two
different Web services, to separate query processing
tasks between basic processing and expansion; (ii)
use of domain ontologies to find alternative ways to
rewrite a query and (iii) adoption of biodiversity stan-
dards to improve sharing and exchanging of informa-
tion on the Web among diferent research groups. On-
tology management is performed by Aondˆe’s Ontol-
ogy Web Service. It provides a wide range of opera-
tions to store, manage, search, rank, analyze and inte-
grate ontologies.
Figure 2 shows a high level view of the Collection
Service and its components. Query processing works
as follows: a Client Aplication sends a user request to
the Collection service (1) with or without request for
expansion. This service encapsulates a Basic Query
module and a Query Expansion service. The Basic
Query module provides a connection with the Collec-
tions Repository, requests query execution (2) and re-
ceives a result (3). If the client did not request query
expansion, the data is returned to the client aplication
(12). Otherwise, the Basic Query module forwards
a request to the Query Expansion service (4). This
service makes a request to our Ontology Web Ser-
vice (5,6) for ontologies that are related to the query.
These ontologies are returned to the Query Expansion
service (7,8) where they are processed to rewrite the
query. The expanded query is sent back to the Basic
Query module (9) which runs it (10) and returns the
result to the client (11,12). The development of these
services is guided by the openness, accessibility and
interoperability provided by open source software and
Web service technologies.
Figure 2: Architecture of the Collection Service.
A query without expansion (Basic query) is a stan-
dard SQL query on the Collections Repository (which
contains field observation and catalog records). The
client application, in such a case, must know the
database schema in order to express the query. The
only acceptable predicates are those that involvefields
that appear in the schema. For instance, the query
“Return all species recorded in the museum catalog
that belong to the family “Amphiuridae” ” will only
work if the database schema has an attribute called
“family”. Table 1 shows an example of a partial re-
sult of this type of query, run on our database.
Table 1: Table with partial query results - basic query.
3.3 The Collections Repository
An important part of our work was the design of the
Collections repository. It is a database containing in-
A TOOL BASED ONWEB SERVICES TO QUERY BIODIVERSITY INFORMATION
307

formation on field obervations and catalog records. It
has been implemented using the PostgreSQL database
system (PostgreSQL, 1996). One relevant issue in
the development of the data model is that it should
be general, allowing the exchange of information bet-
ween different research groups.
For this purpose, we decided to use the data model
elements that are part of the Darwin Core standard
(TDWG, 1994). This means that, in the future, our
work can interoperate with other projects, because it
relies on Web services and in this world wide data
standard. We started by defining the subset of inte-
rest in Darwin Core, and added other relevant specific
fields, specified by our end users.
The entire work was conducted in cooperation
with these end-users: biologists from two distinct re-
search fields - ecology and marine biology. While
the first perform field trips to collect data on inte-
ractions among insects and plants, the latter collect
small sea animals. They are moreover in charge of
a large project to reorganize the university’s zoology
museum, and are thus conversant with the needs and
methods of management of species catalog records.
Thus, our database model reflects a dual view of
biodiversity data management. On one side, we sup-
port storage and handling of data on species obser-
vations and field trip collections. On the other side,
we also cater to the needs of museum catalogs, which
are closer to those of (digital) librarians. As far as we
know, there is no other unifying database model pro-
posal of the same kind - biodiversity databases are ei-
ther concerned with field trip records or with museum
catalog records.
Figure 3 shows a high level view of the database
entity relationship diagram. This multi purpose
database naturally supports a wider spectrum of
queries. This includes for instance queries that trace a
museum record entry back to its field origins, without
losing any of the original annotations.
The central entities of the database model are
Sample (corresponding to field observation/collection
records), HomogeneousSet (records on sets of homo-
geneous species extracted from field collections) and
Catalog (museum records). Sample, Homogeneous
Set and Catalog records have to answer the same kind
of query: What (species identification), How (it was
collected, preserved, catalogued), by Whom, When,
Where. The answer to these queries needs a con-
text (e.g., does the query concern field observations,
catalog entries, or their interconnection). Moreover,
the What (taxonomic information) is often incom-
plete, and may evolve. Location (where) can be er-
roneous or imprecise, when coordinates are unavai-
lable. For more details on data incompleteness in bio-
diversity databases, we refer the reader to (Daltio and
Medeiros, 2008). For more on the collection reposi-
tory, we refer the reader to (Malaverri, 2008).
3.4 The Query Expansion Service
The Collection service receives a query as parameter
and analyzes its predicates and optionally involkesthe
Query Expansion service. The use of ontologies in
query processing allows the Query Expansion service
to expand a query expression to incorporate terms and
concepts that are not in the collection database, but are
part of the biologists’ conceptual view of the world.
This section presents examples of typical queries,
with invocation of the Expansion Service.
3.4.1 The use of Subclasses (Hyponym)
Consider the natural language query:
Return insects of the order lepidoptera that
were collected in the adult life stage.
This query can be represented in SQL (Structured
Query Language) as:
SELECT * FROM Taxonomy t, Catalog
c WHERE t.class=’insecta’ AND t.order =
’lepidoptera’ AND c.lifestage = ’adult’ and
t.idTaxa = c.idTaxa
Suppose the query is posed on Table 2, extracted
from our Catalog Table. In particular, our database
records have many nulls. Hence, records 1, 2 and
3 have the Order identified while 4, 5 and 6 contain
SuperFamily information. The query can be directly
applied to the table, since it contains all needed at-
tributes.
Since the Order attribute is not present directly in
records 4, 5 and 6 these records would not be con-
sidered. However, it is possible to expand the query
using an ontology that represents taxonomic informa-
tion. This ontology is partially depicted in Figure 4.
Using the inheritance relation between the con-
cepts, it is possible to recognize that gracillarioidea,
hesperioidea, micropterigoidea, and papilionoidea
are ontological sub-classes of order lepidoptera. The
query can be rewritten as follows:
SELECT * FROM Taxonomy t, Catalog c
WHERE t.class=’insecta’ AND t.superfamily
in (’Gracillarioidea’, ’Hesperioidea’, ’Mi-
cropterigoidea’, ’Papilionoidea’) AND
c.lifestage = ’adult’ and t.idTaxa = c.idTaxa
The user needs to define whether the query is to be
processed with or without expansion. In the first case,
the query will process only the contents of records 1,
WEBIST 2009 - 5th International Conference on Web Information Systems and Technologies
308
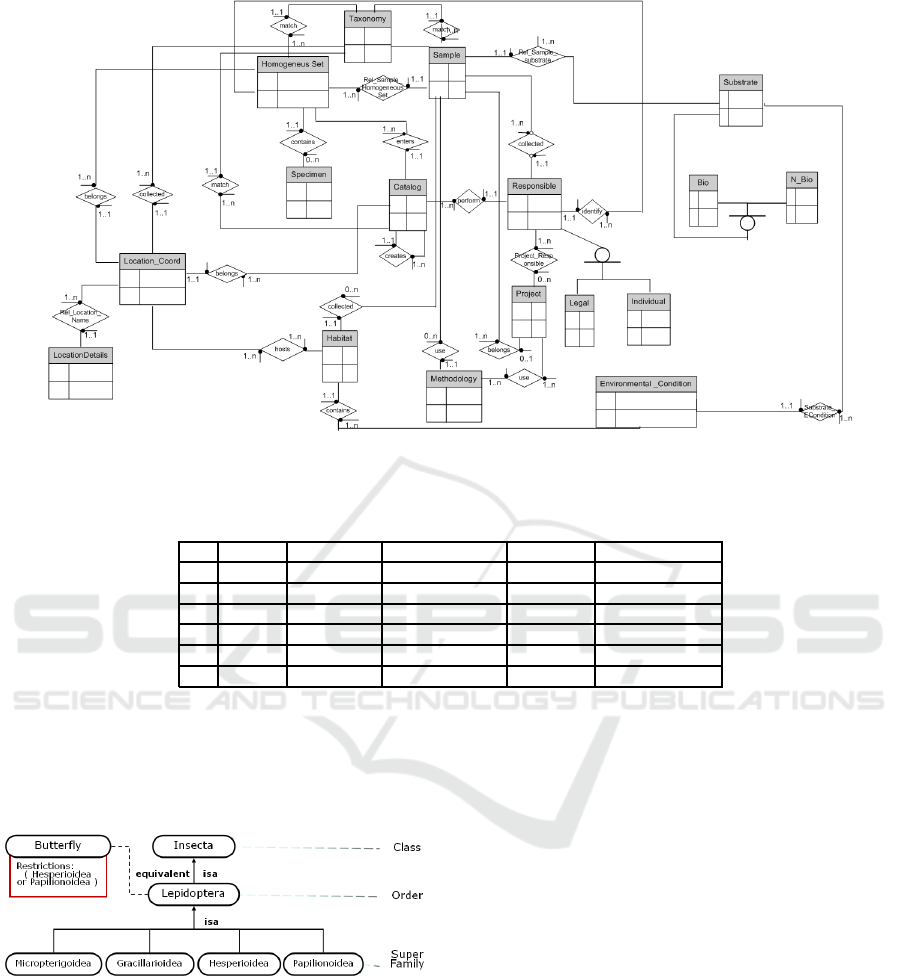
Figure 3: Entity-Relationship Diagram based on Darwin Core standard version 2.
Table 2: Table with records about insects.
Id Class Order SuperFamily LifeStage commomName
1 insecta lepidoptera adult butterfly
2 insecta lepidoptera larval moth
3 insecta coleoptera adult beetle
4 insecta hesperioidea adult
5 insecta lepidotrichidae larval
6 insecta chrysomeloidea adult
2, and 3. In the second case, the Query Expansion ser-
vice is invoked to reformulate the query, and will also
process records 4, 5, and 6. The result is the union of
results of expanded and non-expanded queries.
Figure 4: Partial ontology for the Insecta class.
3.4.2 The use of Equivalence (Synonyms)
Consider the natural language query: ”Return data on
butterflies”. This can be represented in SQL as:
SELECT * FROM Catalog WHERE com-
momName = ’butterfly’
Suppose, again, data on Table 2. If the query is
processed, the criteria specified can be applied di-
rectly only to the records 1, 2 and 3. To consider data
from records 4, 5 and 6, it needs to be reformulated,
e.g., ’common name’ is not present.
Again, it is possible to use the information from
an ontology to specify an alternative classification
mode to verify if an insect is a butterfly. The ontol-
ogy in Figure 4 also includes an alternative concept
that defines this common name, defined equivalent to
order Lepidoptera. However, this equivalence is re-
stricted to Hesperioidea and Papilionoidea (see Fig-
ure 4). From this information, the SQL query can be
reformulated as follows:
SELECT * FROM Catalog WHERE super-
family in (’Hesperioidea’,’Papilionoidea’)
Again, if the user does not demand query expansion,
it will be processed and return record 1. Expansion
will return record 4. The result is the union of both
queries.
3.4.3 Other uses of Ontologies
The previous examples use the information on sub-
class and equivalence relationships to obtain differ-
ent specifications about a concept. Ontologies have
A TOOL BASED ONWEB SERVICES TO QUERY BIODIVERSITY INFORMATION
309

an additional set of resources that can be adopted to
rewrite queries.
Query expansion can consider identity (syn-
tactic identity) and equivalence concepts. Sub-
class/superclass relationships can be exploited in one
or more levels. In particular, super/subclasses can be
suggested to users when a query does not return the
desired answers. Moreover, if a concept consists of
an intersection of others, the query can be specified
utilizing the concepts and restrictions applied to the
intersection.
Additional relationships and properties should be
considered in query expansion. They include applica-
tions of transitivity and symmetry. A particular inter-
esting ontological rewriting possibility involves part
of - whole relationships.
4 CONCLUSIONS
This work presented a tool to support research on bio-
diversity. It uses metadata standards and Web services
to exchange and share data, and applies a query ex-
pansion technique to adapt user queries to the data
sources. Query expansion relies on the use of ontolo-
gies, which are served by a Web service.
The design and test of database and tool are being
conducted with participation of biology experts. The
database has been created using Postgres. The biol-
ogists’ distinct archived files are now being migrated
into the database. We are conducting tests with and
without query expansion, to validate database design
choices. These tests are being executed directly in
SQL. The Collection service has already been speci-
fied and we are now finishing the specification of the
Expansion Service to meet all expansion techniques
of section 3.4.3. All services are being built using
Apache Axis.
Future work involves many issues. The first is
to use the TAPIR protocol, used by large biodiver-
sity projects, as a mechanism to transfer information.
Another issue will involve distinct kinds of user inter-
action modes, and other kinds of interaction mecha-
nisms – e.g., clicking on maps.
ACKNOWLEDGEMENTS
This work was partially financed from grants by
Brazilian funding agencies CNPq (including project
BioCORE), CAPES and FAPESP.
REFERENCES
Andreou, A. (2005). Ontologies and Query expansion.
Master’s thesis, University of Edinburgh.
Beach, J. (2007). Specify Biodiversity Collections Soft-
ware. http://www.specifysoftware.org/Specify/. (ac-
cess Oct, 2008).
Bio-CORE (2008). Tools, models and tech-
niques to support research in biodiversity.
http://www.lis.ic.unicamp.br/projects/bio-core/.
BioCase (2005). Biological Collection Access Services for
Europe. http://www.biocase.org/index.shtml. (access
Oct, 2008).
Colwell, R. (1996). Biota: The Biodiversity Database Man-
ager. Sinauer Associates.
CRIA (2001). Centro de Referˆencia em Informac¸˜ao Ambi-
ental. http://splink.cria.org.br. (access Oct, 2008).
Daltio, J. and Medeiros, C. B. (2008). Aondˆe: An ontol-
ogy web service for interoperability across biodiver-
sity applications. Information Systems, 33. Accepted
for publication.
Florescu, D., Raschid, L., and Valduriez, P. (1996). A
methodology for query reformulation in cis using se-
mantic knowledge. Int. J. Cooperative Inf. Syst.,
5(4):431–468.
GBIF (2004). Global Biodiversity Information Facility.
URL: http://www.gbif.org. (access Oct, 2008).
Gruber, T. (1993). Toward principles for the design for on-
tologies used for knowledge sharing. In Formal On-
tology in Conceptual Analysis and Knowledge Repre-
sentation. Kluwer Academic Publishers.
ITIS (2007). Integrated taxonomic information system.
http://www.itis.gov/. (access Oct, 2008).
Lian, L., Ma, J., Lei, J., Song, L., and Zhang, D. (2007).
Query relaxing based on ontology and users’ behavior
in service discovery. In Fourth International Confer-
ence on Fuzzy Systems and Knowledge Discovery.
Maddison, D. and Schulz, K. (2007). The tree of life web
project. Zootaxa, 1668.
Malaverri, J. G. (2008). Serving Biodiversity Data on the
Web. Master’s thesis. Defence April 2009.
Michener, W., Beach, J., Jones, M., Ludscher, B., Penning-
ton, D., Pereira, R. S., Rajasekar, A., and Schildhauer,
M. (2007). A knowledge environment for the biodi-
versity and ecological sciences. J. Intell. Inf. Syst.,
29(1):111–126.
Necib, C. B. and Freytag, J. C. (2004). Using ontologies
for database query reformulation. In ADBIS (Local
Proceedings).
PostgreSQL (1996). Postgresql.
http://www.postgresql.org/. (access Oct, 2008).
SourceForge.NET (1999). Distributed generic information
retrieval (digir). http://sourceforge.net/projects/digir.
(access Oct, 2008).
TDWG (1994). Biodiversity information standards - tdwg.
http://www.tdwg.org/. (access Oct, 2008).
WEBIST 2009 - 5th International Conference on Web Information Systems and Technologies
310
