
Method for Image Transform Selection in Cytological
Image Analysis
I. Gurevich and I. Koryabkina
Dorodnicyn Computing Centre, Russian Academy of Sciences
40 Vavilov str., Moscow, Russia 119333
Abstract. The paper considers diagnostic analysis of blood system tumours
using special methods. Initial data are images of specimens from patients with
three diagnoses, including two types of aggressive lymphoid tumours, and an
innocent tumour. Analysing feature set, it was found that significant features
vary for different diagnoses. Thus the task requires special methods for image
analysis and recognition, i.e. methods that allow selecting image transformation
depending on informational image nature. The paper shows that applying
special methods, the recognition rate can be increased appreciably.
1 Introduction
The paper considers diagnostic analysis of cytological specimen, in particular blood
system tumours. The distinctive characteristic of the task is that images of different
diagnoses are described by different sets of significant features. Since classical
recognition methods presume that all objects are described with the same feature set
(with possible gaps); the peculiarities of the task cannot be exploited. The task
requires special methods for image analysis and recognition, i.e. methods that allow
selecting image transformation depending on informational image nature.
A method of image transformation selection depending on informational image
nature is applied to solve the task. The method allows taking into account peculiarities
of each class and utilising appropriate recognition algorithms for the objects of each
class. Since the notion of equivalence, which was used in the theoretical background
of the applied method, is originally formulated for algorithms based on estimated
calculations, so we naturally use these algorithms for recognition. It is shown that
recognition rate exceeds 93% for the method.
Section 2 states the set-up of the medical task at hand. It illustrates the pre-
processing stages to form the recognition set, including image enhancement, object
extraction, feature selection and feature calculation. Section 3 explains the
peculiarities of the task and describes the method proposed for task solution. The
steps includes 5 step, followed in detail: 1) Image characterisation, 2) Image model
construction, 3) Definition of equivalence class for image model, 4) Image model
classification, 5) Verification of image characterisation. Section 4 discusses the
results of calculation experiments, shows the difficulties encountered and the
solutions found.
Koryabkina I. and Gurevich I. (2009).
Method for Image Transform Selection in Cytological Image Analysis.
In Proceedings of the 2nd International Workshop on Image Mining Theory and Applications, pages 100-106
DOI: 10.5220/0001964101000106
Copyright
c
SciTePress
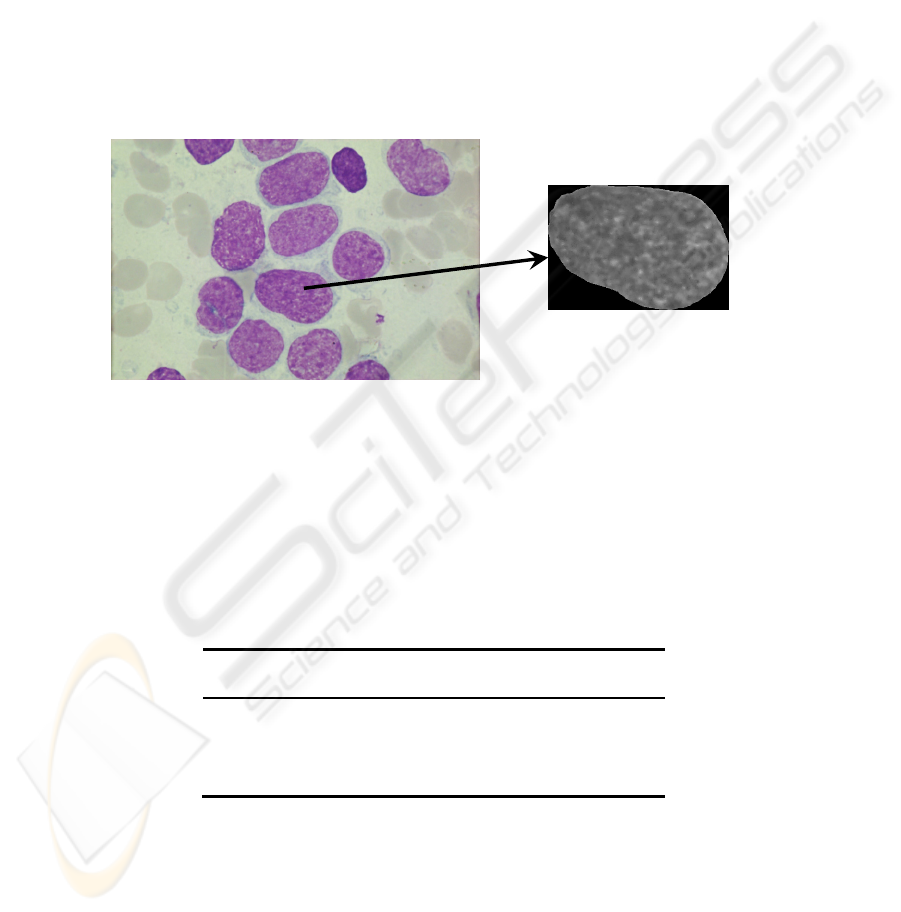
2 Cytological Cell Analysis. Task Set-up
The initial data are images of specimens from patients with three diagnoses, including
two types of aggressive lymphoid tumours: de novo large and mixed cell lymphomas
(CL), and transformed chronic lymphatic leukemia (TCLL), and innocent tumour
(indolent chronic lymphatic leukemia - CLL) [5]. In order to shift from the analysis of
cell images to feature description analysis an information technology is developed in
[2] for morphologic analysis of cytological specimens. Data pre-processing includes
several stages. At the first stage, the medical experts mark diagnostically important
cell nuclei, images of the nuclei are extracted and used for further analysis (see Fig.1).
Fig. 1. Initial data are images of specimens from patients with three diagnoses, including two
types of aggressive lymphoid tumours, and an innocent tumour.
At the second stage, the set of features for nuclei description was formed. In the
process of thorough discussion with medical experts, 47 features were selected,
namely the size of nucleus in pixels, 4 statistical features calculated on the histogram
of nucleus intensity, 16 granulometric and 26 Fourier features of nucleus. The results
of feature measurement form a database, containing diagnostically important
information for 5161 cell nuclei.
Table 1. Initial data.
Diagnosis Patients
(number)
Images
(number)
Nuclei
(number)
CL 18 986 1639
TCLL 12 536 1025
CLL 13 308 2497
Total: 43 1830 5161
The factor analysis is performed on the data set, and the feature sets for each factor
are analysed. Factor analysis shows [3] that for different diagnosis factors of the same
value vary in features with high loads. Consequently, diagnostic value of each feature
varies for different groups of patients. Three groups of diagnostically valuable
features could be distinguished (for feature descriptions see table 2):
101

Features F1, F15, F16 combined with certain features from the range F22 – F29,
1. Feature F2,
2. Features F42, F45.
Table 2. Significant features for cell nuclei description.
Description of the features
F1 Nucleus square in pixels
F2 Mean of intensity histogram
F15 Number of inclusions of typical size
F16 Number of inclusions of minimal size
F22 Mean of F(r)
2
F23 Dispersion of F(r)
F24 The third central moment of F(r)
F25 The forth central moment of F(r)
F26 Number of local maximums of F(r)
F27 Abscissa of global maximum of F(r)
F28 Abscissa of left local maximum of F(r)
F29 Abscissa of right loc. maximum of F(r)
F42
Number of local maximums of F(α)
3
F45
Number of local minimums of F(α)
Thus, each diagnosis can be characterized by the certain number of correlations
between considered features. Analysing factor loads for features from the first and the
second most important factors for different diagnoses, the characteristic sets of
features for each diagnosis can be defined (table 3).
Table 3. Features with high load in factors 1-2 for different diagnoses.
CLL TCLL CL
Factor 1 F2 F22-F29,
F42, F45
F22-F26, F29
F42, F45
Factor 2 F23, F25, F29 F1, F15, F16
Table 3 illustrates that sets of informative features is unique for each diagnoses.
Classical recognition methods presume that each all objects are described with the
same feature set (with possible gaps); the results of factor analysis cannot be
appreciated this case. At the same time, the method for image transformation
selection in recognition tasks [4] supports utilization of these results. The method
allows taking into account peculiarities of each class and utilising appropriate
recognition algorithm for the objects of each class. The method consists of 5 steps,
which are considered below in detail.
2
F(r) is the sum of the elements of Fourier spectrum, that are located on a semicircle with the
center placed in the center of spectrum matrix and radius r.
3
F(a) is the sum of the elements of Fourier spectrum, that are located on the segment starting at
the central element of the spectrum matrix and forming an angle a with the level line
(counter-clockwise).
102

3 Steps of the Applied Method in the Task of Cytological
Specimen Analysis
The section shows how the steps of the method for image transformation selection in
recognition tasks [4] should be adapted for the task at hand. The corresponding
parameters are adjusted for each class, including image equivalence classes,
algorithms for image reduction to recognizable form, image model classes, and
recognition algorithms.
Step 1. Image Characterisation. Image equivalence classes are defined by the
recognition task at hand. Factor analysis shows [2] that feature descriptions of images
for different diagnosis have certain set of correlations, thus certain regularity or
mixture of regularities of different types characterise each class. Consequently, three
equivalence classes {I
1
}, {I
2
}, {I
3
} correspond to the diagnoses, CL, TCLL, and CLL.
Step 2. Image Model Construction. Initial images are described by feature sets.
Therefore variety {T
1
} of algorithms for image reduction to the recognizable form
consists of algorithms for feature calculation (similarly for {T
2
}, and {T
3
}). Note that
feature vectors for image description differ for images from different classes:
{I
1
}: { F2, F23, F25, F29 } ,
{I
2
}: { F22 – F29, F42, F45 } ,
{I
3
}: { F22 – F26, F29, F42, F45 } .
Step 3. Definition of Equivalence Class for Image Model. Since equivalence classes
for image models differ in natural way – feature sets vary for different classes,
equivalence class of image model is determined by the construction. So, by the
construction image model has the same class as the image selected on the first step.
Step 4. Image Model Classification. Notion of equivalence that was used on steps 1
and 2 was originally formulated for algorithms based on estimated calculations
(ACE), so we naturally use ACE for recognition. For experimental study we use
software system «Recognition 1.0» [1], it includes effective implementation of ACE
methods and supports its application for practical task solution. Experiments
demonstrate that the best results are achieved voting by all possible support sets. The
results of recognition are discussed in section 4.
Step 5. Verification of Image Characterisation. At the training stage we naturally
verify the correctness of image characterisation; since we know the correct class when
training, we just compare it with the class obtained. Verification for the recognition
stage is considered in detail at the next section.
It should be emphasized, that since recognition rates vary for the diagnoses, the
sequence of proposing hypothesis becomes essential. The general rule applied here is
as follows: we firstly assume that image belongs to the class with maximum number
of elements, then the second biggest class regarding number of its elements, and so
103
on, and so forth. In this way we decrease the number of calculations and increase
recognition rate.
4 Comparison of Recognition Rates for Different Feature Sets
For experimental purposes objects within each class were arbitrarily divided into two
equal parts, which are training set and recognition set. Recognition rate for the whole
number of features is 86,75% and it varies for the diagnoses (see table 4). High
recognition rate for CLL diagnosis can be explained by the fact that CLL is a non-
malignant disease, while both CL and TCLL are malignant diseases. Therefore, cells
corresponding to CLL diagnosis have pronounced distinctions from the other cells,
while cells of CL and TCLL diagnoses seem to be more similar in appearance.
To test the efficiency of the proposed method, the tests are also performed on the
reduced feature set that includes 14 features determined by factor analysis. The set
contains the following features: { F1, F2, F15, F16, F22 – F29, F42, F45 }. In this
case the recognition rate drops down to 83,18%, but the computational costs also
decrease.
Table 4. Recognition rates for image descriptions consisting of 47 and 14 features.
Diagnosis Total number
(cells)
47 features
1
14 features
CL 820 84,51% 76,34%
TCLL 513 63,35% 58,48%
CLL 1248 97,84% 97,84%
Total 2581 86,75% 83,18%
Now we estimate the recognition rate for the method described in previous section.
To define the parameters of the method an individual training set is constructed for
each equivalence class, it consists of two classes: diagnosis corresponding to the
equivalence class and all the other object marked as “other class”. In other words, for
each class we distinguish the objects of the class from all the other objects. This
necessarily involves the increase in computation time, but should the hypothesis be
properly ordered, the increase is not dramatic.
During computational experiments several major difficulties are encountered and
successfully solved. The first problem is that TCLL diagnosis incorporates only small
number of objects (20% of overall set), and current implementation of ACE is not
efficient in case when classes differ significantly on capacity. So we have to eliminate
certain number of objects from the “other class” in the corresponding set. The set is
cut down to 1547 objects, thus the number of objects of TCLL diagnosis constitutes
not less than 30% of the set (513 objects out of 1547). Recognition rate for TCLL
1
Recognition rate is calculated as ratio between the number of objects attributed to the class
and the number of objects of the class.
104
diagnosis is 96,10%, while only 57,74% of the objects from “other class” are
correctly recognised.
The second difficulty arises for CLL diagnosis. Only one feature (F2) has high load
in the first factor, so support sets cannot be constructed for this case and ACE cannot
be applied. Taking into account that the first factor explains only 21,1% of the set [2],
we decide to take into consideration features that have high load in the second factor
(the second factor for CLL diagnosis explains 17,17% of features), which are features
F23, F25, and F29. The training is performed using the extended feature set, and the
recognition rate for CLL diagnosis is 94,31% (1177 objects out of 1248), for “the
other” diagnosis – 89,20% (1189 objects out of 1333).
For the CL diagnosis 90,24% of object are attributed to the correct class (740
objects out of 820), and 81,94% of object for the “other class” (1443 out of 1761
objects).
Thus it becomes clear that the recognition rate is quite high for each diagnosis and
exceeds 90%. Table 5 summarizes recognition rate for the method applied.
Table 5. Recognition rate for the method.
Diagnosis Correct
recognition
(cells)
Total number
(cells)
Recognition
rate
CL 740 820 90,24 %
TCLL 493 513 96,10 %
CLL 1177 1248 94,31 %
Total 2410 2581 93,37 %
Thus, applying the described method we can raise the recognition rate from 83,18%
up to 93,37%, which is more than 10% increase. This is particularly important for
medical tasks, where patient’s treatment depends on the diagnosis posed. It should be
recorded that recognition rate is higher for the method applied and reduced feature
set, then for the general set of 47 features, which also confirms the efficiency of the
method.
5 Conclusions
Diagnostic analysis of blood system tumours is considered, including data from
patients with three diagnoses (two types of aggressive lymphoid tumours, and an
innocent tumour). The distinctive characteristic of the task is that different feature sets
correspond to different diagnoses. Since classical recognition methods presume that
all objects are described with the same feature set (with possible gaps); the
peculiarities of the task cannot be exploited. This requires special methods for image
analysis and recognition, i.e. methods that allow selecting image transformation
depending on informational image nature.
A method of image transformation selection depending on informational image
nature proved to be efficient for the task, it allows to use different parameters or even
105

different algorithms in order to distinguish the objects of each class. Algorithms based
on estimated calculations are selected for recognition, and their parameters adjusted
for each class. This allows increasing the recognition rate for the task for 10%.
Acknowledgements
This work was partially supported by the Russian Foundation for Basic Research
(Grants Nos. № 08-01-00469, 07-07-13545, 08-01-90022) and by the Program
“Fundamental Sciences for Medicine 2009” of the Presidium of the Russian Academy
of Sciences.
References
1. Yu.I. Zhuravlev, V.V. Ryazanov, O.V. Sen’ko: "RECOGNITION. Mathematical methods.
Software system. Practical Applications", M.: FAZIS (2006).
2. I.B. Gurevich, D.V. Harazishvili, O. Salvetti, A.A. Trykova, I.A. Vorob’ev: Elements of the
Information Technology of Cytological Specimen Analysis: Taxonomy and Factor
Analysis. In: Pattern Recognition and Image Analysis: Advances in Mathematical Theory
and Applications, Vol.16, No.1. MAIK "Nauka/Interperiodica"/Pleiades Publishing, Inc.
(2006) 113-115.
3. I.Gurevich, D.Kharazishvili, D.Murashov, O.Salvetti, I.Vorobjev: Technology for
Automated Morphologic Analysis of Cytological Slides. Methods and Results. In:
Proceedings of the 18th International Conference on Pattern Recognition (ICPR2006),
August 20-24, 2006, Hong Kong, China. The Institute of Electrical and Electronics
Engineers, Inc. (2006) 711-714.
4. I.Koryabkina: Method for Image Informational Properties Exploitation in Pattern
Recognition. In: Proceedings of the 13
th
Scandinavian Conference on Image Analysis, June
– July 2003. J.Bigun and T. Gustavsson (Eds.), SCIA 2003, LNCS 2749 (2003) 1006-1013.
5. Vorob’ev A.I., ed. Atlas “Tumors of Lymphatic System”, Hematological Scientific Center
of the Russian Academy of Medical Sciences (2001).
106
