
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE
KNOWLEDGE RESOURCES
Victoria Nebot and Rafael Berlanga
Departamento de Lenguajes y Sistemas Inform´aticos, Universitat Jaume I
Campus de Riu Sec, 12071, Castell´on, Spain
Keywords:
Ontology generation, Ontology indexing, Knowledge repositories.
Abstract:
Nowadays very large domain knowledge resources are being developed in domains like Biomedicine. Users
and applications can benefit enormously from these repositories in very different tasks, such as visualization,
vocabulary homogenizing and classification. However, due to their large size and lack of formal semantics,
they cannot be properly managed and exploited. Instead, it is necessary to provide small and useful logic-
based ontologies from these large knowledge resource so that they become manageable and the user can take
benefit from the semantics encoded. In this work we present a novel framework for efficiently indexing and
generating ontologies according to the user requirements. Moreover, the generated ontologies are encoded
using OWL logic-based axioms so that ontologies are provided with reasoning capabilities. Such a framework
relies on an interval labeling scheme that efficiently manages the transitive relationships present in the domain
knowledge resources. We have evaluated the proposed framework over the Unified Medical Language System
(UMLS). Results show very good performance and scalability, demonstrating the applicability of the proposed
framework in real scenarios.
1 INTRODUCTION
Ontologies are a knowledge representation formalism
that capture the semantics of the real world entities
by defining concepts and the relationships between
them. The knowledge captured in ontologies can be
used, among other things, to share common under-
standing of the structure of information among people
and/or software agents, to enable the reuse of domain
knowledge and to introduce standards to allow inter-
operability (Noy et al., 2001). Recently, many ap-
plication domains are being represented using ontolo-
gies, one of the most prominent being the life-science
fields. An example of a large repository of biomedi-
cal information is the Unified Medical Language Sys-
tem (UMLS). This knowledge resource is a vast and
very comprehensive repository of information. How-
ever, the large amount of different terminologies it
gathers makes it hard to select a relevant and man-
ageable subset for an specific application. Moreover,
UMLS is not represented in a standard interchange
ontology language such as the W3C recommended
OWL, which prevents the knowledge base from the
benefits of logic-based formalisms.
In this paper, we tackle the evident scaling prob-
lems when dealing with very large and complex
knowledge resources. Our proposal consists of
a framework for building compact and customized
logic-based ontologies from large knowledge re-
sources. The proposed system allows the user to spec-
ify a query as free-text, which encapsulates the se-
mantics the output ontology should gather. The input
query goes through a process of semantic annotation
and later assessment in order to obtain a set of rel-
evant and meaningful ontology concepts. These set
of concepts, called signature, is used by the Ontology
Extractor to obtain a specific application-oriented on-
tology in OWL format. To our best knowledge, our
approach is the first one on addressing the following
requirements:
• Scalability. Knowledge resources tend to be
large and complex repositories of information.
We achieve scalability through the use of tools
aimed at extracting customized ontologies from
the knowledge resources, which just involve the
necessary elements.
• Ontology Compactness. Specific application
ontologies are usually subject-oriented, which
means they gather semantically related concepts.
The compactness of these ontologies depends
greatly on the precision of the semantic annota-
144
Nebot V. and Berlanga R. (2009).
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE KNOWLEDGE RESOURCES.
In Proceedings of the 11th International Conference on Enterprise Information Systems - Artificial Intelligence and Decision Support Systems, pages
144-151
DOI: 10.5220/0001984001440151
Copyright
c
SciTePress
tion of the user query. Therefore, we propose an
assessment step that filters the semantic annota-
tions obtained based on the conceptual density of
each selected ontology concept.
• Logic-based Formalism. The ontologies built
from the knowledge resources are represented in
OWL, which offers several advantages such as
reasoning capabilities, consistency of terminol-
ogy, sharing and reusing knowledge, etc.
The rest of the paper is organized as follows. Sec-
tion 2 describes an application scenario that motivates
the proposed approach of building OWL ontologies
from large knowledgeresources. Section 3 explainsin
detail the different components of the framework and
Section 4 shows the evaluation performed. Finally,
Section 5 gives some conclusions and future work.
2 APPLICATION SCENARIO
In the biomedical domain, the gathering and repre-
sentation of knowledge have been the main concerns
of researchers and domain experts for several years.
Supporting evidence is the great effort made by the
National Library of Medicine (US) in developing the
UMLS over the past 18 years. UMLS can be thought
as a large repository of biological and clinical infor-
mation covering multiple domains: from genetics and
celular to disease level. The version 2007AC has
1,516,299 concepts and 13,226,382 relations. UMLS
includes three resources: the Metathesaurus, the Se-
mantic Network, and the Specialist Lexicon. The first
one consists of several thesauri (more than 100 differ-
ent sources such as FMA, MeSH, etc.) The Seman-
tic Network consists of 135 semantic types organised
in 7 main groups - e.g. anatomical structure or bi-
ologic function - and linked by “is
a” relationships.
There are also 53 kind of non-hierarchical relation-
ships among these semantic types, e.g. causes(Virus,
Disease). A concept of the metathesaurus can be as-
signed at one or more semantic types in the Semantic
Network. The SPECIALIST Lexicon, the third re-
source of UMLS, has been developed in order to pro-
vide lexical information needed for an NLP applica-
tion e.g. syntactic, morphological. It includes many
biomedical terms, dictionaries of abbreviations, etc.
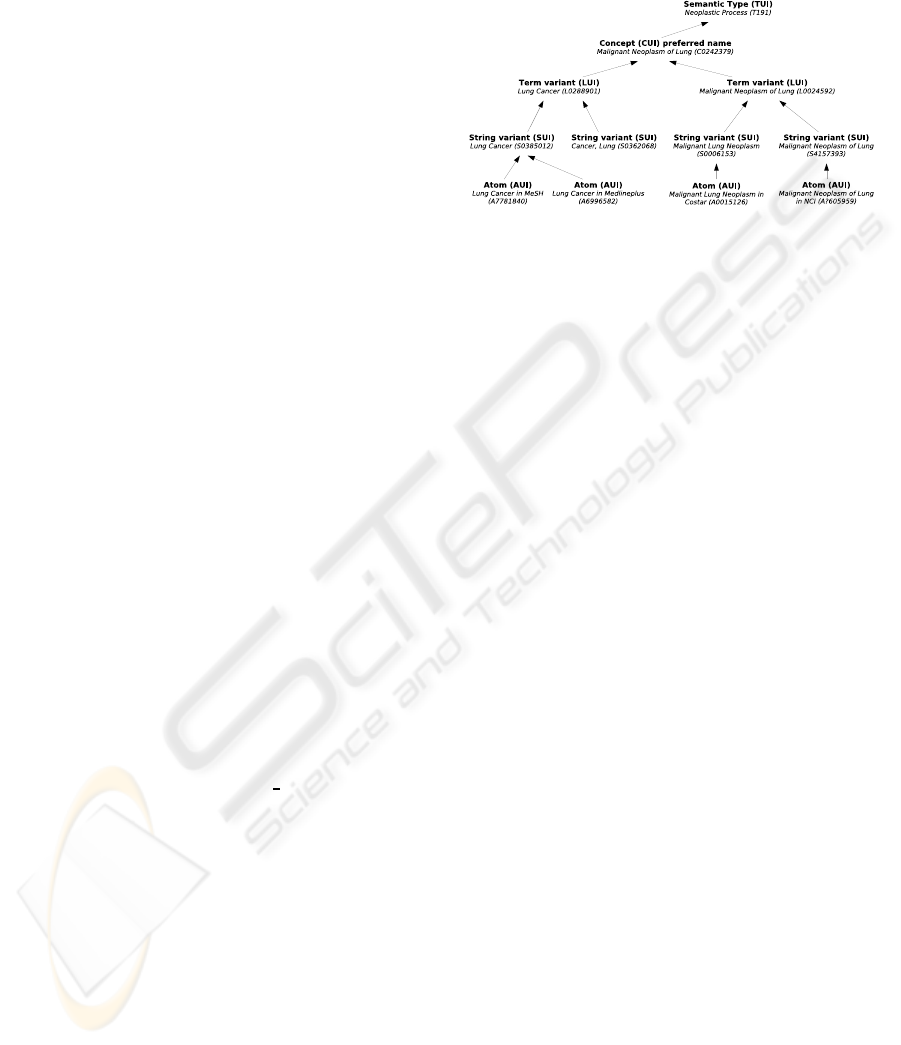
The conceptual model of UMLS is centred on
Sources (i.e. ICD-10, MesH), which contain Atoms
(the distinct concepts within each source). Each Atom
is assigned to one and only one Concept, whose iden-
tifier is called CUI. Concepts consist in Terms ow-
ing the same meaning. One Term can belong to sev-
eral concepts but not often. Terms consist of Strings,
i.e. string variants. Figure 1 shows an example of
the conceptual model of UMLS. The proposed frame-
work works at the level of concept identifiers (CUIs).
Figure 1: Conceptual model of UMLS.
UMLS can be considered a vast and complex on-
tology, from which is hard to select a relevant man-
ageable subset for a specific application. Moreover,
the metathesaurus gathers vocabularies that have been
developed independently, resulting in a lot of incon-
sistencies such as cycles in hierarchies. Moreover,
the UMLS knowledge resource has a proprietary for-
mat, which prevents interoperability and the use of
standard languages and tools designed for the Seman-
tic Web. One could think of translating the whole
metathesaurus into OWL. However, authors such as
(Cornet and Abu-Hanna, 2002) and (Kashyap and
Borgida, 2003) claim this is not feasible since the se-
mantics of the relations present in the vocabularies
(e.g. ICD, MeSH) is often underspecified. Therefore,
UMLS relations can be interpreted in several ways.
As a result, only the Semantic Network has been
translated into OWL (Kashyap and Borgida, 2003).
Our proposal addresses this issue by building specific
ontologies tailored to the needs of a concrete applica-
tion. The output ontology should be used by knowing
that it is a specific interpretation of the metathesaurus.
Moreover, we put special emphasis in building on-
tologies in a logic-based formalism such as OWL due
to the several advantages it offers. OWL brings pow-
erful reasoning based on Description Logic (DL). In
the biomedical domain, reasoning can be a very pow-
erful tool to infer new or implicit knowledge. For ex-
ample, it can serve to make relevant links between two
different biomedical domains. Moreover, we can also
check the consistency of the terminology and perform
more meaningful queries. On the other hand, adding
a description logic to a large coarse-grained concep-
tual model of a biomedical domain serves to disam-
biguate imprecise representations. Finally, by using
OWL, which is a W3C standard, we can benefit from
the tools made available for OWL as well as share and
reuse knowledge in an easy way.
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE KNOWLEDGE RESOURCES
145
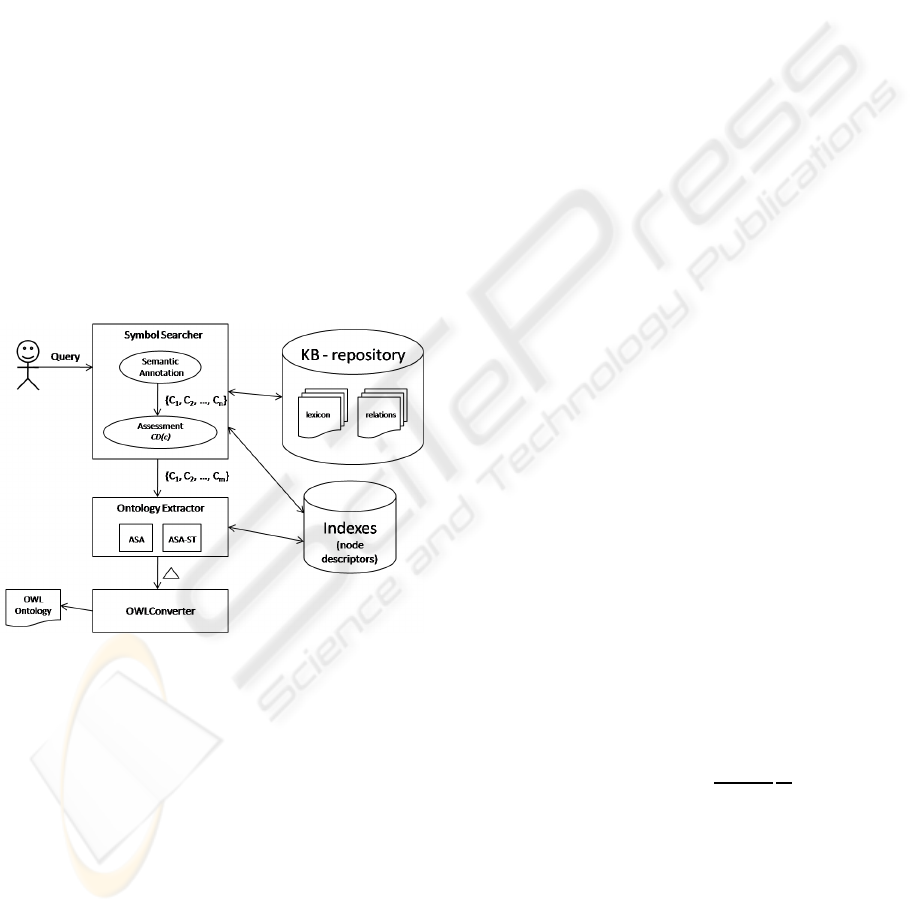
3 SYSTEM ARCHITECTURE
In this section we present the framework for building
tailored application ontologies from very large knowl-
edge resources. Figure 2 shows the different compo-
nents of the framework. The workflow of the frame-
work is as follows: the user specifies a free-text query
that describes the application ontology he wants to
obtain. The Symbol Searcher tool performs the Se-
mantic Annotation of the user query with the lexicon
of the knowledge resource. The set of obtained con-
cepts go through an Assessment step that filters the
concepts according to their conceptual density. The
output of the Symbol Searcher tool is the signature,
which is composed by the knowledge resource con-
cepts that best match the user query both syntacti-
cally and semantically. Taking as input the signature,
the Ontology Extractor retrieves from the knowledge
resource the sub-ontology (necessary concepts along
with their taxonomic relationships) that best covers
the signature. In a final step, the ontology is converted
into OWL format and further enriched with additional
OWL axioms.
Figure 2: Framework for building logic based application
ontologies from knowledge resources.
Next sections are devoted to explain with more de-
tail the main components of the framework, which are
in turn the Symbol Searcher, Ontology Extractor and
OWL Converter.
3.1 Symbol Searcher
The Symbol Searcher is in charge of finding the ap-
propriate set of ontology concepts that best match the
user query. This process is carried out in two phases,
which will be explained in turn: Semantic Annotation
and Assessment.
3.1.1 Semantic Annotation
The main idea behind semantic annotation is the iden-
tification of relevant terms and the linking of these
terms to thesauri, databases or ontologies. In our ap-
plication scenario, we are interested in finding rel-
evant terms in the user query and linking them to
UMLS concepts. One option consists of performing
manual annotation. However, this process is subject
to human errors and factors such as familiarity with
the domain, scale of the knowledge resource, etc. An
alternative to overcome these difficulties is the auto-
matic semantic annotation of text. MetaMap (Aron-
son, 2001) and Whatizit (Rebholz-Schuhmann et al.,
2007) are examples of automatic semantic annotators
that use large terminological databases as input lexi-
cons. MetaMap has been specifically designed to map
text into concepts from the UMLS Metathesaurus
while Whatizit can be used on several resources such
as UniProtKb/Swiss-Prot, GO, UMLS, etc. Both se-
mantic annotators have been tested in the context of
the application scenario and, although they are effec-
tive, a supervised process is still required to complete
and verify the obtained annotations. Since we want all
the steps of the framework to be as automatic as pos-
sible, in the next section we propose an assessment
step that filters out the concepts found by the seman-
tic annotator.
3.1.2 Assessment
The quality and compactness of the resulting applica-
tion ontology is severely affected by the input signa-
ture used to generate it. As earlier mentioned, auto-
matic semantic annotators are subject to imprecission
and ambiguity. Therefore, the objectiveof this assess-
ment step is to evaluate and rank the output ontology
concepts found by the semantic annotator according
to their conceptual density. Let C be the set of con-
cepts found by the semantic annotator and N its size.
We define the conceptual density of a concept of C as
follows:
CD(c) =
N
∑
c
i
∈C,c
i
6=c
1
d(c,c
i
)
1
N
(1)
where d(c, c
i
) is the taxonomic distance between con-
cepts c and c
i
in the knowledge resource.
Notice that CD(c) = 1 if the rest of concepts from
C are direct neighbours of c. Therefore, concepts with
biggerCD are closer one another in the knowledge re-
source, while concepts with smaller CD are far away
from the rest of concepts in C. The latter probably
correspond to incorrect or ambiguous semantic anno-
tations. In order to filter them out the user can specify
ICEIS 2009 - International Conference on Enterprise Information Systems
146

a minimum Threshold as shown in Figure 2. With this
procedure we remove concepts found by the seman-
tic annotator that may introduce noise in the resulting
ontology.
3.2 Ontology Extractor
The Ontology Extractor task consists of retrieving
from the knowledge resource an application ontology
(e.g. a set of concepts along with their taxonomic re-
lationships) that fulfils the user needs. The applica-
tion ontology has to provide the necessary semantics
demanded by the user query while keeping a reduced
size in order to achieve scalability. In order to ac-
complish both requirements we have developed and
tested an indexing mechanism over the “is-a” rela-
tionships of the knowledge resource that enables us to
efficiently retrieve a small subset that satisfies the user
query. Next section describes the indexing process of
the knowledge resource and then we present an ontol-
ogy retrieval strategy based on the indexes created.
3.2.1 Knowledge Resource Indexing Process
In most knowledge resources, as is the case of UMLS,
concepts are organized into “is-a” hierarchies, which
constitute the backbone of the repository. This leads
to an underlying graph-like structure. In order to effi-
ciently retrieve a sub-ontology from this graph struc-
ture guided by the signature, we need some kind of
indexing scheme over the graph that encodes descen-
dant and ancestor relationships in a compressed and
efficient way. We have adopted a labeling scheme,
which assigns to each node in the graph some iden-
tifier that allows the computation of relationships
between nodes using simple arithmetic operations.
For our purposes, we have adopted and extended
Agrawal’s interval scheme (Agrawal et al., 1989) but
with a labeling variation from (Schubert et al., 1983),
which takes preorder identifiers of nodes instead of
postorders used in Agrawal’s technique. The ap-
proach can be applied to directed trees and Directed
Acyclic Graphs (DAGs), which will be the underly-
ing structure of most ontologies (Christophides et al.,
2003). With respect to our application scenario, we
have preprocessed UMLS in order to delete cycles in
the “is-a” hierarchy and obtain a DAG.
Figure 3 (left graph) shows a labeled DAG. The
process is as follows: in an initial step, disjoint com-
ponents can be hooked together by creating a virtual
root node. The compression scheme first finds a span-
ning tree T for the given graph (solid edges). Then it
assigns an interval to each node based on the preorder
traversal of T. That is, the interval associated with a
node v is [pre(v), maxpre(v)], where pre(v) is the pre-
order number of v and maxpre(v) is the highest pre-
order number of v’s descendants. Notice the preorder
of each node is used as its unique identifier. Next, all
nodes of the graph are examined in the reverse topo-
logical order so that for every edge from node p to
q, all the intervals associated with node q are added
to the intervals associated with node p, taking into
account that if one interval is subsumed by another,
the subsumed interval is not added. In the figure, the
interval [2, 7] is associated to node d when labeling
the spanning tree. Then, during the reverse topologi-
cal traversal, node d inherits intervals [6, 6], [4, 6] and
[5, 5] corresponding to nodes g, e and h, which come
from the dashed edges not belonging to the spanning
tree. Since these intervals are already subsumed by
d’s interval [2,7], they are not added to d. Otherwise,
they would be included.
The storage requirements for trees labeled with
this interval scheme is O(n), since one interval per
node is enough. For DAGs, the worst case requires
O(n
2
) space. However, this situation is unlikely be-
cause Agrawal’s approach for DAGs finds the opti-
mum spanning tree, that is, the spanning tree that
leads to minimum amount of intervals per node and
thus, minimum storage requirements.
Next step consists of obtaining analogous infor-
mation about ancestors of each node. The strategy ap-
plied is as follows. First, we reverse the edges of the
original structure so that each node now points to its
parent/s (see right graph of Figure 3). Then, a virtual
root node has to be created to hook together what are
leaf nodes in the original structure. Then, the same la-
beling scheme described previously is applied to the
reversed structure. Since now the edges denote an-
cestor relationships, the labeling scheme will encode
ancestor nodes. Notice that each node identifier is its
preorder number and both the original structure and
the reversed one have each own preorder system.
We finally define the descriptor function of a node
v as follows:
descriptor(v) =<descpre(v), descintervals(v),
ancpre(v), ancintervals(v),
topo(v) >
where descpre(v) denotes the preorder number of v in
the original structure, descintervals(v) denotes the set
of intervals encoding v’s descendants, ancpre(v) de-
notes the preorder number of v in the reversed struc-
ture, ancintervals(v) denotes the set of intervals en-
coding v’s ancestors and topo(v) denotes the topolog-
ical order of v.
Gathering all together, we have designed an en-
coding mechanism for concepts in a knowledge re-
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE KNOWLEDGE RESOURCES
147
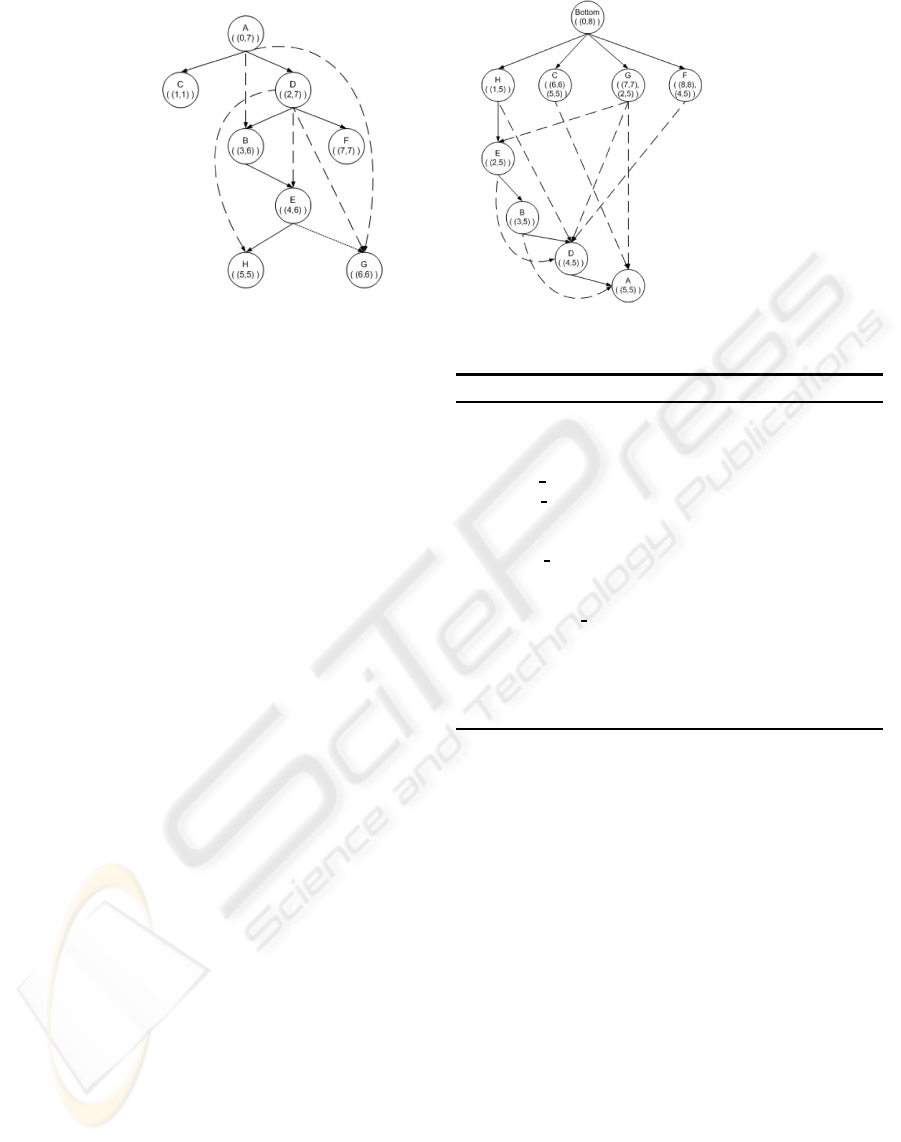
Figure 3: Compressed transitive closure of descendants (left graph) and ancestors (right graph).
source based on interval labeling schemes that is able
to encapsulate in a compressed and efficient way all
the descendants and ancestors of each concept. The
information encapsulated in the concept descriptors
can be exploited to efficiently retrieve related con-
cepts. Therefore, we define a set of operations in-
tended to manipulate the intervals of the descriptors.
The most important operations are the next:
• The descendants of v is the serialization of
descintervals(v).
• The ancestors of v is the serialization of
ancintervals(v).
• The topological order of v is topo(v).
• Concept v
1
subsumes v
2
if:
descintervals(v
1
) ∩ descintervals(v
2
) ==
descintervals(v
2
)
• Common ancestors of v
1
and v
2
are:
ancintervals(v
1
) ∩ ancintervals(v
2
).
• Nearest common ancestor of v
1
and v
2
is:
max{topo(commonAncestors(v
1
, v
2
))}.
3.2.2 Ontology Retrieval Strategy
The aim of the strategy presented in this section is to
extract ontologies with a reduced size and relevant to
the user query. The output constitutes the skeleton
of the application ontology (“is-a” relationships with
tree-like structure). This ontology skeleton can be
further enriched with additional “is-a” relationships
in order to obtain a DAG structure, which has the
property of preserving all the taxonomic relationships
among the input signature concepts. This section is
dedicated to describe the strategy followed in order
to achieve such a goal.
Algorithm 1: Compute spanning tree.
Require: L list of output nodes sorted by preorder number
Ensure: G spanning tree of the output nodes
Stack parents =
/
0
C = next node(L)
D = next node(L)
while L do
if subsumes(descintervals(C), descintervals(D) then
add edge(G, edge(C, D))
push(parents, D)
C = D
D = next node(L)
else
pop(parents)
C = top(parents)
end if
end while
All Signature Ancestors (ASA). The ontology re-
trieval strategy presented consists of extracting all an-
cestors from the signature concepts encoded in the de-
scriptors. Then, Algorithm 1 computes a spanning
tree with the signature and their ancestors based on
their subsumption relationships encoded in the de-
scendant intervals. The output ontology contains all
concepts from the signature plus all their ancestors
organized by their “is-a” relationships.
Figure 4 shows an example of the output ontolo-
gies (tree-like structure) that would be retrieved by
this strategy and the refinement presented next tak-
ing as starting point the same signature. Figure 4.a
shows the underlying graph-like “is-a” relationships
of the original knowledge resource, in which thicker
lines correspond to the spanning tree calculated by the
interval labeling scheme described in Section 3.2.1.
Figure 4.b including the crossed out nodes corre-
sponds to the ontology skeleton that would be re-
trieved by ASA strategy, in which black nodes are the
input signature.
ICEIS 2009 - International Conference on Enterprise Information Systems
148
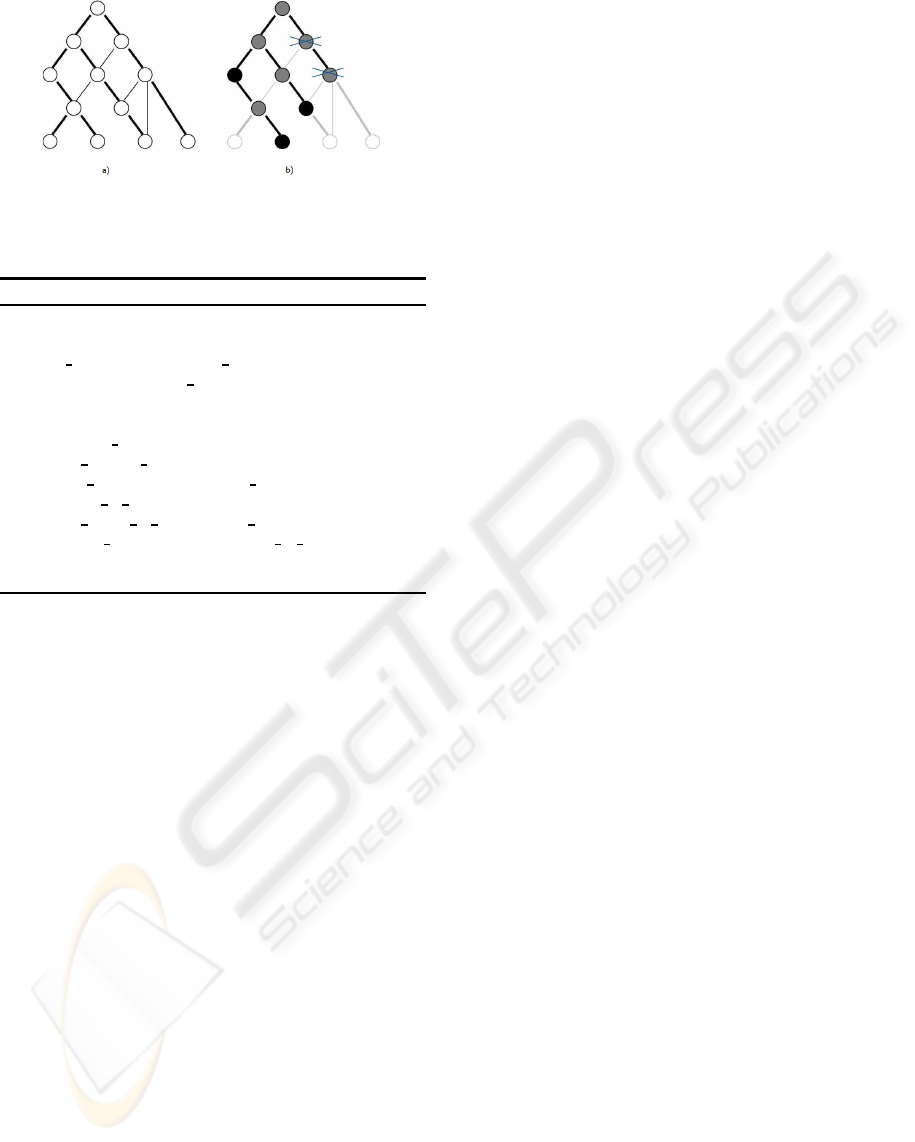
Figure 4: Output ontology skeleton. a) Original knowledge
resource “is-a” relationships; b) Output ontology of ASA
(all nodes) and ASA-ST (crossed out nodes not included).
Algorithm 2: Search additional edges to get a DAG.
Require: G output tree structure of ASA or ASA-ST
Ensure: G with DAG-structure
sorted nodes = topological order(G)
for all node n in sorted nodes do
ancestors = ancestors(n)
while ancestors do
nearest ancestor =
get nearest ancestor(n, ancestors)
add edge(G, edge(nearest ancestor, n)
nodes to root =
get nodes to root(nearest ancestor)
delete from(ancestors, nodes to root)
end while
end for
All Signature Ancestors Spanning Tree (ASA-ST).
In the previous approach, retrieving all ancestors
from the signature concepts can result in an excessive
amount of nodes in the output ontology that are not
relevant to the reconstructed hierarchy. Thus, we
have introduced an enhancement by just selecting
ancestor nodes that relate concepts of the signature
through the spanning tree calculated (see crossed
out nodes of Figure 4.b). This approach can be
considered an extension of ASA strategy, since it is
applied to the ASA output ontology. In this strategy,
deleted nodes do not participate in the transitive
closure of the signature, thus, it is not altered. The
experiments performed show that ASA-ST strategy
is more adequate when working with UMLS due
the great amount of unrelevant ancestors pruned.
Therefore, we take ASA-ST strategy as the one
suitable for our application scenario.
Obtaining a DAG. The output ontology consists of a
tree structure for simplicity, since many applications
do not require all “is-a” relationships among concepts
but rather a tree hierarchy (e.g. datawarehouse dimen-
sions). However, in many cases it is necessary to get
the complete DAG structure. For this purpose, the Al-
gorithm 2 has been designed.
This algorithm adds the remaining relationships
between pairs of nodes belonging to the signature.
This is accomplished by calculating nearest ancestors
using the topological order of nodes.
This algorithm does not add an extra temporal
complexity because all operations have a linear cost
w.r.t. the number of nodes if they make use of the
descriptor functions of each node.
3.2.3 UMLS Specific Customizations
The indexing scheme and ontology retrieval strategy
presented in this paper are not specific to UMLS.
They can be applied to any knowledge resource with
some kind of transitiverelationship (e.g. “is-a”, “part-
of”, etc.). However, once the ontology skeleton is ob-
tained one can enrich it with specific features from the
knowledge resource. In the case of UMLS, concepts
belong to one or more semantic types as explained
in Section 2. Therefore, we enrich the graph structure
obtained in the previous phase with the corresponding
semantic types and “is-a” relationships.
3.3 OWL Converter
The final step performed by the system consists of
building an interchangeable OWL file with the graph-
based structure returned by the Ontology Extractor.
Table 1 summarizes the patterns applied to the graph
to obtain the final OWL ontology. For the sake of
space, we use DL notation instead of OWL construc-
tors. Axiom 1 converts a node, which represents a
concept, into an OWL class. Then, axiom 2 adds a
subClassOf axiom for each edge, which represents an
“is-a” relationship. By applying the previous axioms
we obtain a taxonomy where classes are primitive, i.e.
without explicit definition. In order to enrich the on-
tology with reasoning capabilities, we need to make
explicit some concept definitions using existing ones.
Axioms 3 and 4 performthis task. In particular,axiom
3 defines a concept as the intersection of its parents,
while axiom 4 defines a concept as the union of its
children. Additionally, axiom 5 includes UMLS re-
lationships as ObjectProperties and Restrictions over
the involved concepts only if both domain and range
are included in the graph so that we obtain closed on-
tologies. Following (Kashyap and Borgida, 2003), we
only add axiom 5 if the corresponding relationship is
not blocked in the Semantic Network. Finally, we can
further enrich the ontology concepts and properties
with annotations provided by the knowledge resource
(e.g. label, comment, user-defined annotation proper-
ties, etc).
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE KNOWLEDGE RESOURCES
149
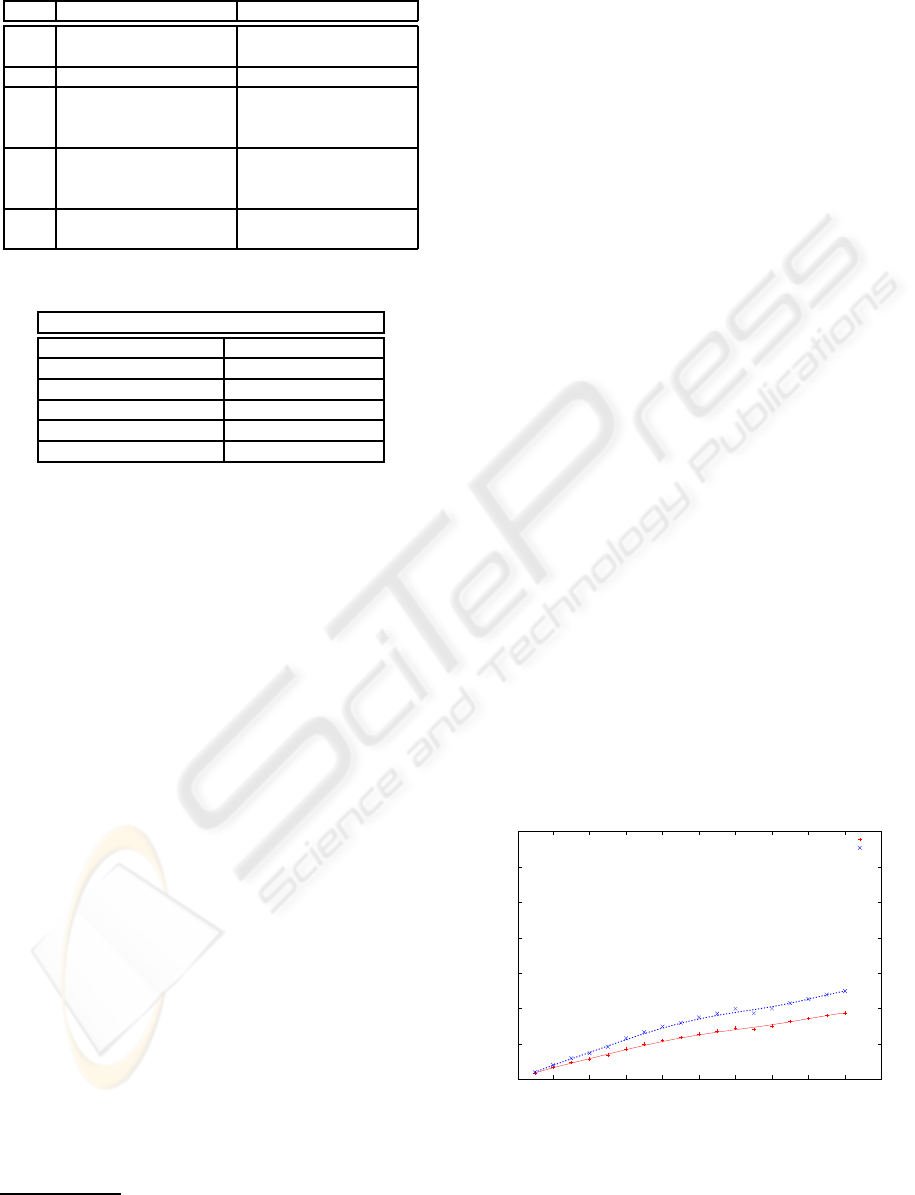
Table 1: OWL conversions from a DAG structure.
DAG OWL (DL)
(1) node(C) C
C ∈ CUIs∪Semtypes
(2) edge(D,C) C ⊑ D
(3) edge(D
1
,C)
. . . C ⊑ (≡)D
1
⊓ . . . ⊓ D
n
edge(D
n
,C)
(4) edge(C, D
1
)
. . . C ≡ D
1
⊔ . . . ⊔ D
n
edge(C, D
n
)
(5) node(A), node(C) A ⊑ ∃R.C
R(A,C)
Table 2: UMLS statistics: # intervals per descriptor.
UMLS number of intervals per descriptor
Number of descriptors 293041
Avg. descendants 2.28
Max. descendants 2326
Avg. ancestors 11.46
Max. ancestors 114
Depth 27
4 EVALUATION
In the following, we describe the experiments per-
formed over the UMLS to prove that our framework
is both scalable and able to retrieve relatively small
sized ontologies compared to the whole knowledge
resource. A prototype was implemented in Python
and MySql has been used as back-end storage system
for the indexes.
Firstly, Table 2 shows some statistics about the
size of the indexes generated by our ontology index-
ing system (concept descriptors) for UMLS. As we
can observe, the maximum number of intervals in a
concept descriptor can be quite large but this is not
usual at all since the average in the descendant inter-
vals is really low while in the ancestors’ increases but
not too much. Thus, we can state that our indexes
are scalable when dealing with large knowledge re-
sources. Some of these repositories have a high de-
gree of dynamism, since they are continously evolv-
ing and growing(e.g. a new version of UMLS is made
available every few months, more or less). This is not
quite a problem for our framework since newer ver-
sions of repositories can be indexed in a few minutes.
The performance evaluation of the framework has
been carried out over a set of 100 different signatures
derived from a collection of 15000 abstracts taken
from MEDLINE. These signatures have been devel-
oped in the context of the Health-e-Child
1
project,
1
Health-e-Child Project: http://www.health-e-child.org
where each signature represents a different perspec-
tive for the study of a given disease. We selected
the diseases Juvenile Idiopathic Arthritis (JIA) and
Tetralogy of Fallot (TOF). Abstracts are processed
with a semantic tagger, in our case (Jimeno et al.,
2008), which identifies UMLS concepts from texts.
These concepts are grouped according to the target
disease (JIA or TOF) and their semantic type. For
the assesment phase, the conceptual density has been
shown quite useful to detect both wrongly annotated
strings and non-relevant concepts derived from the
user’s request. In the experiments, we have com-
bined the conceptual density with the frequency in
which concept strings occur in the collection. In this
way, concepts with high frequency and low density
are likely to be wrong annotations, whereas concepts
with low frequency and relative low density are not
relevant to the signature. In both cases, concepts are
removed from the signature. Notice that this reduc-
tion contributes to the increase of both the compact-
ness and the performance of the ontology construction
process. In our experiments the density threshold is
around 0.1, under which concepts can be potentially
rejected. The resulting groups constitute the signa-
tures for the experiments.
Figure 5 shows the size of the output ontology as
the size of the signature increases for both versions
of strategy ASA-ST (tree and DAG). The size of the
ontology (axis Y) is measured in number of edges
since both versions extract an ontology with the same
amount of nodes. The increase of edges in the DAG
version is due to the fact that UMLS mixes different
classifications over the same concepts, and this leads
to multiple inheritance. Overall, we can predict a lin-
ear tendency of the ontology size as the signature in-
creases in both versions.
0
200
400
600
800
1000
1200
1400
20 40 60 80 100 120 140 160 180 200
# of edges
signature’s size
Increase of edges from spanning tree to DAG with ASA-ST
spanning tree
DAG
Figure 5: Signature’s size vs. ontology’s size.
We have also evaluated the temporal complexity
of both ASA-ST versions (see Figure 6). Although
ICEIS 2009 - International Conference on Enterprise Information Systems
150
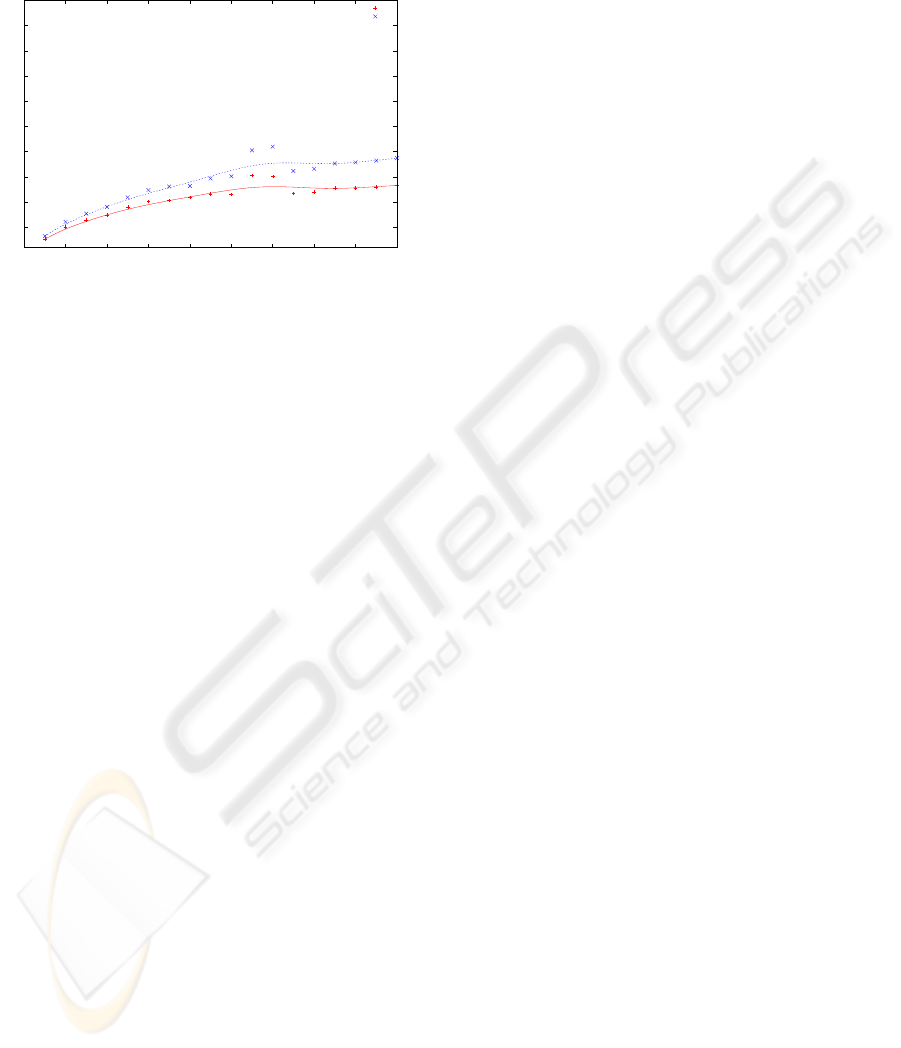
the time required to generate a DAG is slightly larger,
the general tendency is linear in both cases.
5
10
15
20
25
30
35
40
45
50
0 20 40 60 80 100 120 140 160 180
time (s)
signature’s size
Signature’s size vs. time
ASA-ST (tree)
ASA-ST (DAG)
Figure 6: Signature’s size vs. temporal complexity.
5 CONCLUSIONS
Using semantics and managing efficiently very large
domain knowledge resources suffers severely from
scaling problems. The framework presented in this
paper is a way of overcoming these difficulties. De-
velopers can use it to quickly create an application
logic-based ontology that covers just a small part of
the knowledge resource. Moreover, ontologies gener-
ated and enriched with a DAG-structure preserve the
taxonomic relationships over the signature concepts.
Evaluation has shown ontologies generated with
this framework keep their size relatively small and
manageable according to their signature. There-
fore, the framework presented achieves scalability by
providing compact and logic-based OWL ontologies
from very large knowledge resources.
In the future, we plan to study different relevance
judgements for the assessment phase that help filter-
ing out semantic annotations. We are also interested
in finding an automatic method for the calculation of a
threshold in this phase. Other future lines focus more
on the relationships of the knowledge resource and in
finding efficient indexing techniques that allow to ef-
ficiently manage them.
REFERENCES
Agrawal, R., Borgida, A., and Jagadish, H. V. (1989). Ef-
ficient management of transitive relationships in large
data and knowledge bases. In SIGMOD ’89: Proceed-
ings of the 1989 ACM SIGMOD international confer-
ence on Management of data, pages 253–262, New
York, NY, USA. ACM.
Aronson, A. R. (2001). Effective mapping of biomedical
text to the UMLS metathesaurus: the metamap pro-
gram. Proc AMIA Symp, pages 17–21.
Christophides, V., Plexousakis, D., Scholl, M., and Tourtou-
nis, S. (2003). On labeling schemes for the semantic
web. In WWW, pages 544–555.
Cornet, R. and Abu-Hanna, A. (2002). Usability of expres-
sive description logics – a case study in UMLS. In
Proc. AMIA Symp, pages 180–4.
Jimeno, A., Jimenez-Ruiz, E., Lee, V., Gaudan, S.,
Berlanga, R., and Rebholz-Schuhmann, D. (2008).
Assessment of disease named entity recognition on a
corpus of annotated sentences. BMC Bioinformatics,
9(Suppl 3):S3.
Kashyap, V. and Borgida, A. (2003). Representing the
UMLS semantic network using owl: (or ”what’s in a
semantic web link?”). In Fensel, D., Sycara, K. P., and
Mylopoulos, J., editors, International Semantic Web
Conference, volume 2870 of Lecture Notes in Com-
puter Science, pages 1–16. Springer.
Noy, N. F., Sintek, M., Decker, S., Crub´ezy, M., Fergerson,
R. W., and Musen, M. A. (2001). Creating seman-
tic web contents with prot´eg´e-2000. IEEE Intelligent
Systems, 16(2):60–71.
Rebholz-Schuhmann, D., Arregui, M., Gaudan, S., Kirsch,
H., and Yepes, A. J. (2007). Text processing through
web services: Calling whatizit. Bioinformatics, pages
btm557+.
Schubert, L. K., Papalaskaris, M. A., and Taugher, J.
(1983). Determining type, part, color and time rela-
tionships. IEEE Computer, 16(10):53–60.
BUILDING TAILORED ONTOLOGIES FROM VERY LARGE KNOWLEDGE RESOURCES
151
