
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL
PROCESSES
Ermelinda Oro
DEIS, University of Calabria, Via P. Bucci 41/C, Rende (CS), Italy
Massimo Ruffolo
ICAR-CNR, University of Calabria, Via P. Bucci 41/C, Rende (CS), Italy
Keywords:
Knowledge Representation and Reasoning, Knowledge Management, Ontology, Workflow, Data Mining,
Workflow Mining, Decision Support System, Health Care Information System, Clinical Process, Medical
Knowledge.
Abstract:
Managing costs and risks is an high priority theme for health care professionals and providers. A promising
approach for reducing costs and risks, and enhancing patient safety, is the definition of process-oriented clini-
cal information systems. In the area of health care information systems, a number of systems and approaches
to medical knowledge and clinical processes representation and management are available. But no systems
that provide integrated approaches to both declarative and procedural medical knowledge are currently avail-
able. In this work a clinical process management system aimed at supporting a semantic process-centered
vision of health care practices is described. The system is founded on an ontology-based clinical knowledge
representation framework that allows representing and managing, in a unified way, both medical knowledge
and clinical processes. The system provides functionalities for: (i) designing clinical processes by exploit-
ing already existing and ad-hoc medical ontologies and guideline base; (ii) executing clinical processes and
monitoring their evolution by adopting alerting techniques that aid to prevent risks and errors; (iii) analyzing
clinical processes by semantic querying and data mining techniques for making available decision support
features able to contain risks and to enhance cost control and patient safety.
1 INTRODUCTION
Across the world the issue of patient safety, medical
errors preventionand healthcare risk management is a
very challenging and widely studied research and de-
velopment topic. It stimulates a growing interest in
the computer science researchers community.
A promising approach for reducing errors and
risks, and enhancing patient safety, is the definition
of process-oriented clinical information systems. In
fact, healthcare services and practices are character-
ized by complex clinical processes in which high risk
activities take place. A clinical process can be seen
as a workflow where clinical (e.g. treatments, drugs
administration, guidelines execution, medical exam-
inations) and general (e.g. patient enrolment, medi-
cal record instantiation, risk evaluation) activities and
events occur. Clinical processes and their activities
are, also, characterized by specific and sophisticated
medical knowledge. Systems that provide integrated
functionalities for representing and managing medi-
cal knowledge and for designing, executing (taking
into account risks rules and conditions) and analyzing
clinical processes, can change clinical practices and
help diffusion of a process and quality awareness in
healthcare organizations.
Currently, as described in Section 2, in the field
of healthcare information systems, a number of ap-
proaches to medical knowledge and guidelines repre-
sentation and management have been proposed. Ex-
isting systems and approaches suffer of the following
shortcomings: (i) they have a lack of mechanisms for
errors and risks handling and prevention; (ii) they do
not use the same formalism for representing and man-
aging both medical knowledge and clinical processes,
hence, they are not able to exploit in a fully unified
way declarative and procedural knowledge during ex-
ecution and monitoring of clinical process; (iii) they
do not allow to organize clinical processes and their
element as an ontology; (iv) they do not allow to mod-
ify and customize represented knowledge and to exe-
cute clinical process in a flexible and agile way.
180
Oro E. and Ruffolo M. (2009).
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL PROCESSES.
In Proceedings of the 11th International Conference on Enterprise Information Systems - Artificial Intelligence and Decision Support Systems, pages
180-187
DOI: 10.5220/0001994401800187
Copyright
c
SciTePress

This work describes the prototypical implemen-
tation of a Clinical Process Management System
(CPMS) that aims at supporting a semantic process-
centered vision of health care practices. The system
is founded on an Ontology-based Clinical Knowledge
Representation Framework (OCKRF) that allows to
express in a combined way medical ontologies, clini-
cal processes and errors and risks rules. More in de-
tail, the OCKRF allows the CPMS to provide meth-
ods for: (i) creating ontologies of clinical processes
that can be queried and explored in a semantic fash-
ion; (ii) expressing errors and risks rules (by means
of reasoning tasks) that can be used (during processes
execution) for monitoring processes; (iii) executing
clinical processes and acquiring clinical process in-
stances by means of either workflow enactment (pre-
defined process schemas are automatically executed)
or workflow composition (activity to execute are cho-
sen step-by-step by humans); (iv) monitoring clini-
cal processes during the execution by running reason-
ing tasks; (v) analyzing acquired clinical process in-
stances, by means of querying and inference capabili-
ties, in order to recognize errors and risks for patients.
The CPMS adopts a semantics approach for rep-
resenting and managing both static and dynamic as-
pects of medical knowledge. So it enables better de-
sign, execution, control and management of clinical
processes and related errors and risks rules. CPMS
delivers health care professionals semantic decision
support functionalities able to contain risks (due to
medical errors and adverse events) in order to enhance
patient safety.
The remainder of this paper is organized as fol-
lows. Section 2 describes related work in the field of
healthcare information systems. Section 3 sketches
the OCKRF which the CPMS is founded on. Section
4 depicts system features and architecture by using an
application example. Finally, Section 4 concludes the
paper and sketches future work directions.
2 RELATED WORK
In the recent past, a strong research effort has been
taken to provide standard representations of declara-
tive and procedural medical knowledge. In the fol-
lowing, available approaches and systems for medi-
cal ontologies and clinical process representation and
management are described.
Medical knowledge representation area provides
one of the most rich collection of domain ontolo-
gies available worldwide. A very famous and widely
adopted thesaurus is Mesh the Medical Subject Head-
ings classification (MESH). It provides a controlled
vocabulary in the fields of medicine, nursing, den-
tistry, veterinary medicine, etc. MeSH is used to in-
dex, catalogue and retrieve the world’s medical liter-
ature contained in PubMed. Another classification,
that has become the international standard diagnos-
tic classification for all medical activities and health
management purposes, is ICD10-CM (ICD; WHO)
the International Classification of Diseases Clinical
Modification, arrived to its 10th Revision. The most
comprehensive medical terminology developed to
date is SNOMED-CT (SNOMED), the Systematized
Nomenclature of Medicine Clinical Terms, based on
a semantic network containing a controlled vocabu-
lary. Electronic transmission and storing of medi-
cal knowledge is facilitated by LOINC, the Logical
Observation Identifiers Names and Codes (LOINC),
that consists in a set of codes and names describ-
ing terms related to clinical laboratory results, test
results and other clinical observations. Machine-
readable nomenclature for medical procedures and
services performed by physicians are descried in CPT,
the Current Procedural Terminology (CPT), a regis-
tered trademark of the American Medical Associa-
tion. A comprehensive meta-thesaurus of biomedi-
cal terminology is the NCI-EVS (NCI-EVS) cancer
ontology. Some medical ontologies are, also, due
to European medical organizations. For example,
CCAM the Classification Commune des Actes Med-
icaux (CCAM), is a French coding system of clinical
procedures that consists in a multi-hierarchical classi-
fication of medical terms related to physician and den-
tal surgeon procedures. A classification of the termi-
nology related to surgical operations and procedures
that may be carried out on a patient is OPCS4, the Of-
fice of Population Censuses and Surveys Classifica-
tion of Surgical Operations and Procedures 4th Revi-
sion (OPCS-4), developed in UK by NHS. The most
famous and used ontology in the field of healthcare
information systems is UMLS, the Unified Medical
Language System (UMLS), that consists in a meta-
thesaurus and a semantic network with lexical appli-
cations. UMLS includes a large number of national
and internationalvocabulariesand classifications (like
SNOMED, ICD-10-CM, and MeSH) and provides a
mapping structure between them. This amount of
ontologies constitutes machine-processable medical
knowledge that can be used for creating semantically-
aware health care information systems.
The evidence-based medicine movement, that
aims at providing standardized clinical guidelines for
treating diseases (Sackett et al., 1996), has stim-
ulated the definition of a wide set of approaches
and languages for representing clinical processes. A
well known formalisms is GLIF, the Guideline Inter-
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL PROCESSES
181

change Format (GLIF). It is a specification consist-
ing of an object-oriented model that allows to repre-
sent sharable computer-interpretable and executable
guidelines. In GLIF3 specification is possible to re-
fer to patient data items defined by a standard med-
ical vocabularies (such as UMLS), but no inference
mechanisms are provided. Proforma (Sutton and Fox,
2003) is essentially a first-order logic formalism ex-
tended to support decision making and plan execu-
tion. Arden Syntax (Pryor and Hripcsak, 1993; Peleg
et al., 2001; HL7) allows to encode procedural med-
ical knowledge in a knowledge base that contains so
called Medical Logic Modules (MLMs). An MLM is
a hybrid between a production rule (i.e. an ”if-then”
rule) and a procedural formalism. It is less declarative
than GLIF and Proforma, its intrinsic procedural na-
ture hinders knowledge sharing. EON (Musen et al.,
1996) is a formalism in which a guideline model is
represented as a set of scenarios, action steps, deci-
sions, branches, synchronization nodes connected by
a ”followed-by” relation. EON allows to associate
conditional goals (e.g. if patient is diabetic, the tar-
get blood pressures are 135/80) with guidelines and
subguidelines. Encoding of EON guidelines is done
by Protg-2000 (Prot´eg´e) knowledge-engineering en-
vironment.
The group of formalisms, presented above, aim
at representing either static (ontologies) or procedural
(guidelines) medical knowledge. They pay less or no
attention to the combined specification and manage-
ment of both static and procedural aspects of medi-
cal knowledge. Furthermore, they do not hold ad-hoc
mechanisms that allow errors and adverse events pre-
vention. In fact, no reasoning facilities able to ex-
ploit patient and disease status, prescribed cares and
drugs, current activities to execute, etc. are provided.
This limitations hinder clinical processes monitoring
aimed at discovering situations that could create risks
for patients during processes execution.
Two interesting healthcare information systems
that provide comprehensive framework for managing
clinical guidelines are DeGeL (Shahar et al., 2003)
and SEBASTIAN (Kawamoto and Lobach, 2005).
DeGeL is focused on providing automated support
for the specification and implementation of clini-
cal guidelines in the treatment of patients, particu-
larly those with chronic conditions such as diabetes,
hypertension and depression. SEBASTIAN (Sys-
tem for Evidence-Based Advice through Simultane-
ous Transaction with an Intelligent Agent across a
Network) captures medical knowledge in XML doc-
uments known as Executable Knowledge Modules
(EKMs). An EKM encapsulates medical knowledge
in a machine-executable format that can be used to
generate patient-specific inferences useful for clinical
decision support (CDS). EKMs use a patient infor-
mation model based on the HL7 Reference Informa-
tion Model (HL7) , and medical concepts are prefer-
entially defined using standard vocabularies included
in UMLS. These systems are mainly designed to sup-
port decisions during diagnosis or guidelines appli-
cation but do not support complex reasoning over
available knowledge for risk management scopes. A
further remarkable system is ASTI (S´eroussi et al.,
2001) because it is the only existing system that tack-
les the problem of errors prevention in prescriptions.
The ASTI project, in fact, has been focused on the
design of a guideline-based decision support system
to help general practitioners avoid prescription errors
and comply with best therapeutic practice, specifi-
cally in the treatment of chronic diseases including
hypertension.
From the above discussion of related works
emerges that already existing approaches and sys-
tems: (i) have a lack of mechanisms for errors and
risks handling and prevention; (ii) do not use the same
formalism for representing and managing both medi-
cal knowledge and clinical processes, hence, they are
not able to exploit in a fully unified way declarative
and procedural knowledge during execution and mon-
itoring of clinical process; (iii) do not allow to orga-
nize clinical processes and their element as an ontol-
ogy in order to provide semantic querying and brows-
ing capabilities; (iv) do not allow to modify and cus-
tomize represented knowledge and to execute clinical
process in a flexible and agile way. The system de-
scribed in the following section aims at overwhelm
these limitations.
3 THE FRAMEWORK
The Ontology-based Clinical Knowledge Represen-
tation Framework (OCKRF) allows to represent
machine-executable and flexible model of declarative
(static) and procedural(dynamic) medical knowledge.
The framework is organized in tree layers as shown in
Figure 1.
The first layer, called OCKRF meta-model is
founded on an ontology-based approach to medical
knowledge representation that allows to express in a
combined way medical ontologies and clinical pro-
cesses. The adopted approach is grounded on a meta-
model that merges expressive power of ontology and
workflow representation formalisms. More in detail,
the meta-model is expressed by means of a formal-
ism based on an object-oriented version of datalog
that holds typical ontology representation constructs
ICEIS 2009 - International Conference on Enterprise Information Systems
182
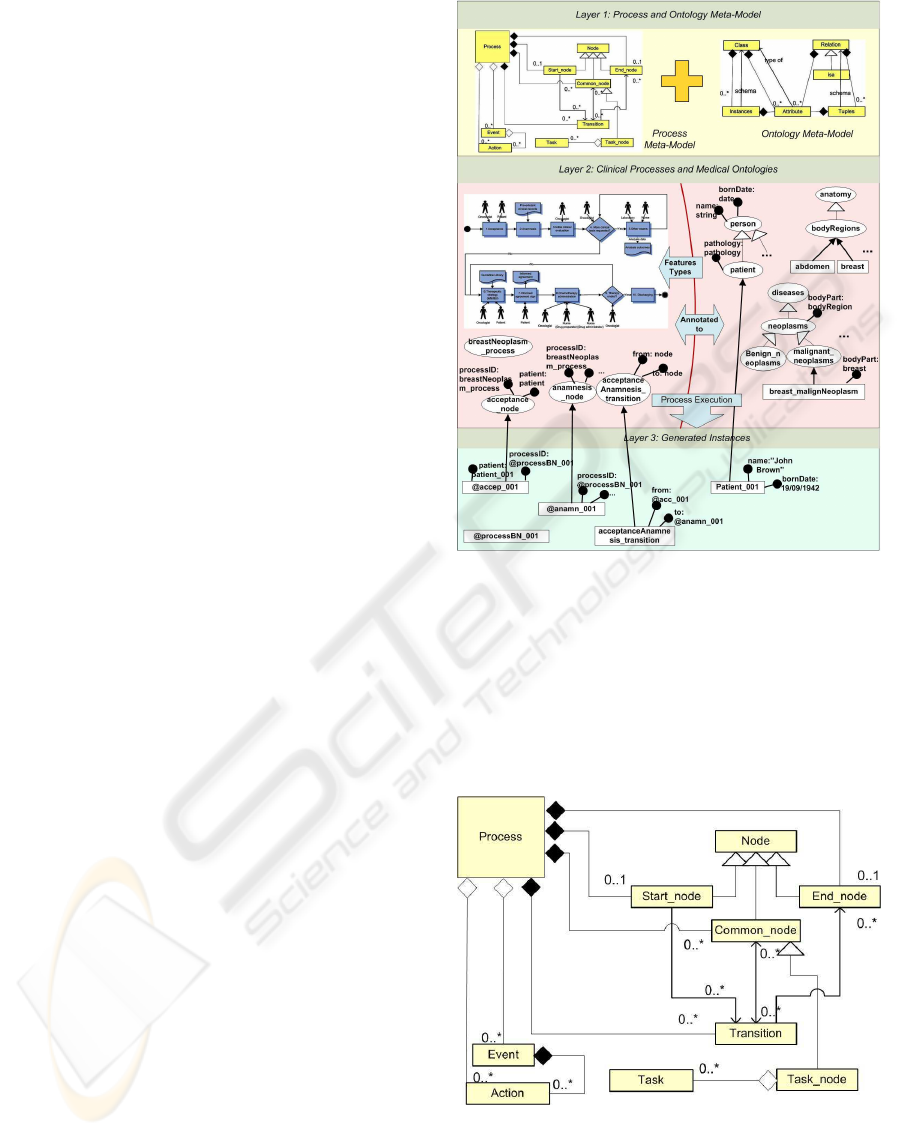
like: classes, relations, attributes, objects (instances).
Furthermore, querying and reasoning capabilities of
Datalog allow to query represented ontologies and ex-
ecute reasoning task over them in a semantic fash-
ion. The meta-model exploits a flow-graph oriented
workflow modeling approach (Figures 2 and 3) in-
spired to the JPDL (JPDL). In a flow-graph based
approach a workflow is represented by a labeled di-
rected graph whose nodes correspond to the activities
to be performed, and whose arcs describe the prece-
dences among them. The key idea, which the pre-
sented framework is based on, is that elements of the
workflow meta-model (i.e. processes, nodes, tasks,
events, transitions, actions, decisions) are expressed
as ontology classes. So, by using the adopted ap-
proach ontologies of clinical processes can be ob-
tained. So, by using the adopted approach: (i) clin-
ical processes and their elements can be organized as
an ontology (for instance, possible events can be de-
fined as a taxonomy in which each class of event has a
specific set of attributes); (ii) workflow elements like
nodes, taskes, activities, events etc. are represented
in terms of ontology classes so types of their param-
eters can be other classes, hence clinical process can
be managed in a semantic fashion; (iii) decisions can
be defined as reasoning tasks that involve not only the
execution state but also the knowledge represented in
the medical ontologies (for example, at each prescrip-
tion a reasoning task can check if the administered
drug is compatible with allergies and/or with the state
of the patient); (iv) special reasoning tasks aimed at
controlling possible risks and errors conditions can be
defined for a process (reasoning tasks generate events
that are properly handled in the process).
The second layer is constituted by medical ontolo-
gies and clinical processes expressed by the formal-
ism provided in the first layer. In particular: (i) med-
ical ontologies represent concepts related to different
medical domains (e.g. diseases, drugs and their inter-
actions, prescription criteria, medical examinations,
medical treatments, laboratory terms, anatomy, pa-
tients administration concepts, possible errors). Med-
ical ontologies can be obtained by importing already
existing ontologies and thesaurus or designed by hand
(for example, concepts regarding patients, wards,
hospital, could depend from the specific hospital or
ward). For instance, concepts related to breast neo-
plasm, needed in the process presented in the Sec-
tion 4.1, has been imported from the International
Classification of Diseases (ICD10-CM), the Anatom-
ical Therapeutic Chemical (ATC) classification sys-
tem, and the Medical Subject Headings (Mesh) Tree
Structures. (ii) clinical processes are both medical
guidelines and clinical practices depending from the
Figure 1: The Ontology-based Clinical Knowledge Repre-
sentation Framework.
specific ward. Clinical processes can by imported
from already available guideline bases or designed by
hand when they represent clinical practices followed
in a specific ward for caring a given disease. Medi-
cal ontologies and clinical processes are stored in the
system knowledge base.
Figure 2: The process meta-model.
The third layer consists of a set of medical ontolo-
gies and clinical process instances stored in a knowl-
edge base. Instances are generated during medical on-
tologies definition and/or clinical process execution.
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL PROCESSES
183
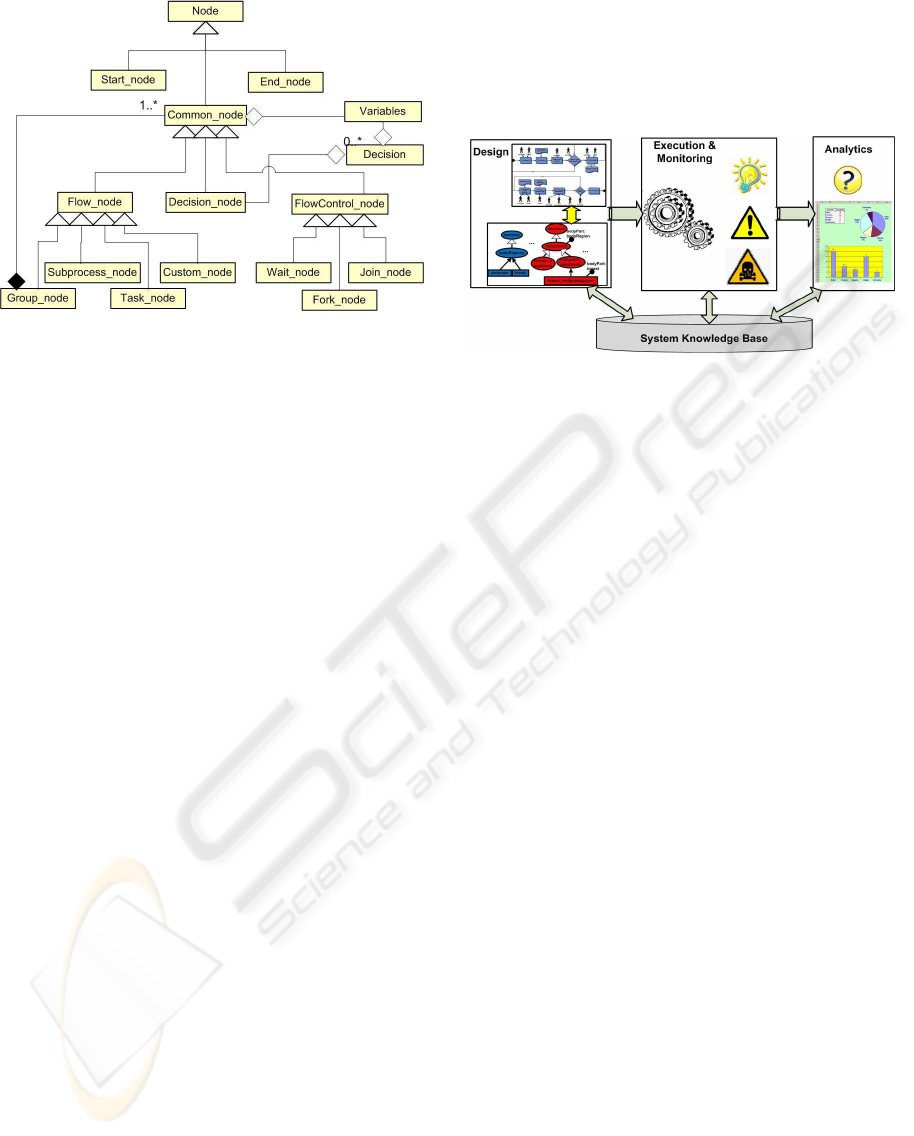
Figure 3: The nodes hierarchy.
Instances are stored in the knowledge base and made
available for querying and reasoning.
More in general, the OCKRF allows methods for:
(i) creating ontologies of clinical processes that can
be queried and explored in a semantic fashion; (ii) ex-
pressing errors and risks rules (by means of reason-
ing tasks) that can be used (during processes execu-
tion) for monitoring processes; (iii) monitoring clin-
ical processes during the execution by running rea-
soning tasks that enable to prevent errors and adverse
events that can cause risks for patients; (iv) to define
semantic Electronic Medical Records schemas (i.e.
Meta EMR); (v) analyzing acquired clinical process
instances, by means of querying and inference ca-
pabilities, in order to recognize errors and risks for
patients. For example, advanced browsing capabili-
ties of the medical knowledge base by using concept-
based queries that enables to retrieve clinical pro-
cesses and guidelines by using available medical on-
tology concepts.
The OCKRF is implemented by the DLP+ ontol-
ogy language (Ricca and Leone, 2007) that beside
complete and expressive ontology representation fea-
tures, holds also powerful ASP reasoning capabili-
ties (Eiter et al., 1997; Leone et al., 2006; Ricca and
Leone, 2007) over represented knowledge.
4 SYSTEM DESCRIPTION
In this section are described architecture (Figure 4)
and functionalities of the the Clinical Process Man-
agement System (CPMS) prototypical implementa-
tion. The prototype has been obtained by combin-
ing the JBPM engine (JPDL) and the DLV+ system
(Ricca and Leone, 2007). The prototype is designed
to follow a clinical processes life-cycle model based
on 3 phases: (i) representing (importing) medical
clinical processes and ontologies; (ii) executing and
monitoring clinical processes; (iii) acquiring, query-
ing and analyzing clinical process instances. Each
phase is implemented by an ad-hoc software module
as described in the following.
Figure 4: The CPMS architecture.
4.1 Clinical Processes Design
The Design module mainly exploits the DLV+ sys-
tem. It provides functionalities for defining medical
ontologies and clinical processes by: (i) import fa-
cilities that allow to acquire already existing ontolo-
gies and guidelines; (ii) direct ”on-screen” drawing
and manual specification functionalities based on an
ontology and process editor. The editor enables ag-
ile guidelines representation and browsing. It con-
tains a rules editor that allows to define ontology con-
straints and/or reasoning tasks used to control risks
and errors during processes execution. Process ele-
ments (e.g. nodes, tasks, decisions) that do not belong
to a specific process can be, also, represented. Ac-
quired and/or represented schemas and instances are
stored in a knowledge base and can be queried by us-
ing querying and meta-querying capabilities of DLV+
system.
In order to briefly describe how the design module
works, in the following is presented, an application of
the system to a real case concerning a clinical process
for caring the breast neoplasm. The clinical process
(depicted in Figure 5) is referred to the practices car-
ried out in the oncological ward of an Italian hospi-
tal, for this reason it is not a general guideline but a
specific clinical process adopted in the domain of the
considered ward.
The clinical process is organized in the following
10 activities and sub-processes:
1. The task node acceptance models the patient en-
rollment. The patient arrives to the ward with an
already existing clinical diagnosis of a breast neo-
plasm. This node is manually executed by an on-
cologist that collect patient personal data that are
ICEIS 2009 - International Conference on Enterprise Information Systems
184

Figure 5: A clinical process for caring the breast neoplasm.
stored as new instances of the classes that describe
process and patient information.
2. The group node anamnesis represents a set of
anamnesis activities that can be executed without
a specific order. Activities in the group are: (i)
general anamnesis, in which physiological gen-
eral data (e.g. allergies, intolerances) are be-
ing collected; (ii) remote pathological anamnesis,
concerning past pathologies; (iii) recent patholog-
ical anamnesis, in which each data or result de-
rived from examinations concerning the current
pathology (or pathologies) are acquired.
3. The task node initial clinical evaluation allows to
acquire the result of an examination of the patient
by a local oncologist.
4. The decision node more clinical test requested
represents the decision of the physician about the
necessity to perform or not additional examina-
tion on the patient.
5. the group node other exams models possible ad-
ditional clinical tests. Each node of the group
correspond an exam. If requested these tests are
conducted to find out general (or particular) con-
ditions of patient and disease not fully deducible
from the test results already available.
6. The task node therapeutic strategy definition
models guideline to follow for caring the given
neoplasm. At design time the physician picks a
guideline (among those available in the knowl-
edge base) that depends upon actual pathology
state as well as other collected patient data. The
semantic indexing strategy allows the retrieval of
guidelines by using concept-based queries. At
each guideline corresponds the prescription of re-
lated drugs. So, the selection of a guideline im-
plies computation of doses, which may depend on
patient’s biomedical parameters, such as body’s
weight or skin’s surface. Cross-checking doses at
execution time (by ad hoc reasoning tasks) is fun-
damental here, because if a wrong dose is given to
the patient the outcome could be lethal.
7. The task node informed agreement sign models
the agreement of the patient concerning under-
standing and acceptance of consequences (either
side effectsor benefits) which may derivefrom the
chosen chemotherapy, and privacy agreements.
8. The sub-process node chemotherapy administra-
tion, models the guideline to execute for caring
the patient.
9. The decision node therapy ended models a control
about effect of the therapy and the possibility to
stop, continue or change cares.
10. The task node discharging models the discharging
of the patient from the ward and allow to acquire
final clinical parameter values.
By using the rule editor, for each clinical process
(or its elements) a set of risk and error conditions can
be described in terms of ontology constraints and/or
reasoning tasks. So during both manual and auto-
matic executionof clinical processes, these conditions
can be executed in order to check possible risks and
errors that are going to happen. If a condition is veri-
fied, the system generates an event that alerts the actor
that is executing the activity, so risks and errors can be
prevented.
4.2 Clinical Processes Execution
and Monitoring
The Execution & Monitoring module provides func-
tionalities for the assisted execution of clinical pro-
cesses and the acquisition of process instances. The
module is mainly constituted by the JBPM engine that
interact with DLV+ system.
Clinical process execution is performed in two
ways: (i) by using a workflow enactment strategy. In
this case, a process schema, designed and stored in
DLV+, is imported in JBPM and automatically ex-
ecuted involving actors that can be humans or ma-
chines (e.g. legacy systems supplying results of med-
ical examinations); (ii) by using a dynamic workflow
composition strategy. In this case, nodes to execute
are selected step by step by choosing the most ap-
propriate one in a given moment. Nodes are chosen
by using semantic querying capabilities of DLV+ sys-
tem and executed by JBPM. Queries allow to spec-
ify patient clinical data and each significant informa-
tion available in medical ontologies (e.g. drug inter-
action, allergies, etc). So, queries exploit patient clin-
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL PROCESSES
185

ical data coming from anamnesis and medical exam-
ination, and each information available in the partic-
ular moment within ontologies and already executed
process activities. The execution of each activity is
performed either: (i) automatically by executing cus-
tom nodes that run java code designed to involveother
existing systems (e.g. batch acquisition of clinical re-
sults from a computer posted in a laboratory); or (ii)
manually by a web based graphical user interface that
shows forms to human process actors. During manual
execution physician and nurses can store values of ac-
tivity parameters by filling forms by hand. They can
insert the values by writing them in the related textual
fields. Special queries and reasoning tasks can be trig-
gered to user input in order to check the correctness of
entered values. The execution generates both process
and ontology instances that are stored in the DLV+
knowledge base. Since, process elements (e.g. nodes,
decisions, transitions, tasks) are represented by means
of ontology classes, process instances are in turn con-
stituted by ontology class instances. This way process
schemas and instances intrinsically constitute an on-
tology. Ad hoc extensions permit the interaction be-
tween JBPM and DLV+. So reasoning, querying and
meta-querying over schemas and available instances
are possible.
Clinical process monitoring is based on the auto-
matic execution and verification, on live clinical pro-
cesses, of ontology constraints and reasoning tasks
modeled at design time. This way, the system can
generate events that inform the user about exceptions
and unusual or undesired behaviors. The availabil-
ity of domain medical ontologies concerning drugs’
interactions, side effects and contraindications could
reduce dramatically the probability of fatal mistakes.
Furthermore, the set of activities executed until a
given moment, the current state of a patient, etc. can
be retrieved and visualized by physician and nurse in
order to check the evolution of cares.
By considering the example introduced in the pre-
vious paragraph, at execution time, for each node of
the clinical process and for each ontology concept in-
volved in the process, instances are created. For ex-
ample results of the anamnesis and the check-up are
stored as instances of the noticed pathologies where
attributes are filled with the related observed values.
During guideline execution, due to drug inherent tox-
icity, before each dose administration a check about
drug administration is performed by analyzing pro-
cess evolution, patient conditions, and information
about the particular drug contained in the drug ontol-
ogy (e.g. maximum absolute ratings for certain drug
in a ”whole life” or other ratings relative to biomed-
ical parameters of the patient). Problems related to
drugs administration can be immediately flagged as
dangerous, or even lethal, and then associated risk is
notified to the oncologist.
4.3 Clinical Processes Analytics
The Analytics module aims at allowing analysis of
the clinical processes instances after their acquisi-
tion. The execution of clinical processes, in fact,
makes available process instances organized as an on-
tology. This way a large amount of semantically en-
riched data becomes available for querying and re-
trieval. Analysis are possible by reports composed by
using semantic querying capabilities of DLV+ applied
to process instances contained in the system knowl-
edge base.
5 CONCLUSIONS AND FUTURE
WORK
This work describes the prototypical implementa-
tion of a semantic clinical process management sys-
tem founded on an ontology-based clinical knowl-
edge representation framework. The system allows to
jointly represent both medical knowledge (by means
of medical ontologies) and clinical processes (by
means of a ontology-based workflow representation
approach). The system allows: (i) creating ontolo-
gies of clinical processes that can be queried and ex-
plored in a semantic fashion; (ii) expressing errors
and risks rules (by means of reasoning tasks) that
can be used (during processes execution) for mon-
itoring processes; (iii) manual process execution in
which each clinical activity to perform in a given
moment is chosen by physician on the base of the
current configuration of patient and disease parame-
ters; (iv) automatic execution by means of the enact-
ment of a designed process schema; (vi) automatic
monitoring of clinical processes execution by running
ad-hoc reasoning tasks that exploit knowledge repre-
sented in medical ontologies and clinical processes
to check possible error and risk causes. The execu-
tion of clinical processes allows to acquire process
and ontology instances that are stored in a knowledge
base. Acquired instances can be analyzed by means
of reports obtained by querying the system knowl-
edge base. Currently the system has been applied to
the clinical process depicted in Figure 5. The practi-
cal application shown that the system enables better
health care decision making capabilities that allows
health care professionals to improve risks and errors
prevention.
ICEIS 2009 - International Conference on Enterprise Information Systems
186

The main challenging future research and devel-
opment problems are the definition of an efficient
query engine working on the conjunct representation
of workflows and ontologies and the definition of
further monitoring and analytical technics. In par-
ticular, existing process instances can be organized
in datasets that can be analyzed by means of data
and workflowmining techniquesaimed at discovering
patterns related to risks and adverse events. In partic-
ular, workflow mining techniques are able to classify
clinical process instances on the base of their behav-
ior and, possibly, to suggest new schemas (precess re-
engineering) able to reduce risks for patients and the
impact of errors. Furthermore, new import/exportfea-
tures will be implemented in order to make the system
compliant with already existing health care informa-
tion systems standards.
REFERENCES
CCAM. Classification Commune des Actes M´edicaux
(CCAM). http://www.ccam.sante.fr/.
CPT. Current Procedural Terminology (CPT): A
Registered trademark of the American Med-
ical Association (AMA). http://www.ama-
assn.org/ama/pub/category/3113.html.
Eiter, T., Gottlob, G., and Mannila, H. (1997). Disjunctive
Datalog. ACM Transactions on Database Systems,
22(3):364–418.
GLIF. Guide Line Interchange Format (GLIF). http://
www.glif.org.
HL7. Health Level Seven (HL7). http://www.hl7.org/.
ICD. International Classification of Diseases, Tenth
Revision, Clinical Modification (ICD-10-CM).
http://www.cdc.gov/nchs/about/otheract/icd9/icd10cm.htm.
JPDL. jBPM Process Definition Language (jPDL). http://
www.jboss.org/jbossjbpm/jpdl/.
Kawamoto, K. and Lobach, D. F. (2005). Design, imple-
mentation, use, and preliminary evaluation of sebas-
tian, a standards-based web service for clinical deci-
sion support. In AMIA Symp., pages 380–4.
Leone, N., Pfeifer, G., Faber, W., Eiter, T., Gottlob, G.,
Perri, S., and Scarcello, F. (2006). The dlv system for
knowledge representation and reasoning. ACM Trans.
Comput. Logic, 7(3):499–562.
LOINC. Logical Observation Identifiers Names and Codes
(LOINC). http://loinc.org/.
MESH. Medical Subject headings (MeSH). http://
www.nlm.nih.gov/mesh/2008/index.html.
Musen, M. A., Tu, S. W., Das, A. K., and Shahar, Y. (1996).
Eon: A component-based approach to automation of
protocol-directed therapy. Journal of the American
Medical Informatics Association, 3(6):367–388.
NCI-EVS. NCI Enterprise Vocabulary Services (EVS).
http://ncicb.nci.nih.gov/NCICB/infrastructure/
cacore
overview/vocabulary.
OPCS-4. The Office of Population Censuses and
Surveys Classification of Surgical Opera-
tions and Procedures, 4th Revision (OPCS-4).
http://www.openclinical.org/medTermOPCS4.html.
Peleg, M., Ogunyemi, O., Tu, S., Boxwala, A. A., Zeng, Q.,
Greenes, R. A., and Shortliffe, E. H. (2001). Using
features of arden syntax with object-oriented medical
data models for guideline modeling. In in American
Medical Informatics Association (AMIA) Annual Sym-
posium, 2001, pages 523–527.
Prot´eg´e. Ontology Editor and Knowledge Acquisition Sys-
tem. http://protege.stanford.edu.
Pryor, T. A. and Hripcsak, G. (1993). The arden syntax for
medical logic modules. Journal of Clinical Monitor-
ing and Computing, 10(4):215–224.
Ricca, F. and Leone, N. (2007). Disjunctive logic program-
ming with types and objects: The dlv+ system. J. Ap-
plied Logic, 5(3):545–573.
Sackett, D. L., Rosenberg, W. M. C., Gray, M. J. A.,
Haynes, B. R., and Richardson, S. W. (1996). Ev-
idence based medicine: what it is and what it isn’t.
BMJ, 312(7023):71–72.
S´eroussi, B., Bouaud, J., Dr´eau, H., Falcoff, H., Riou, C.,
Joubert, M., Simon, C., Simon, G., and Venot, A.
(2001). Asti: A guideline-based drug-ordering system
for primary care. MEDINFO.
Shahar, Y., Young, O., Shalom, E., Mayaffit, A.,
Moskovitch, R., Hessing, A., and Galperin, M.
(2003). Degel: A hybrid, multiple-ontology frame-
work for specification and retrieval of clinical guide-
lines. In AIME, pages 122–131.
SNOMED. Systematized Nomenclature of
Medicine-Clinical Terms (SNOMED CT).
http://www.ihtsdo.org/snomed-ct/.
Sutton, D. R. and Fox, J. (2003). The syntax and semantics
of the proforma guideline modeling language. JAMIA,
10(5):433–443.
UMLS. Unified medical Language System (UMLS). http://
www.nlm.nih.gov/research/umls/.
WHO. World Health Organization (WHO). http://
www.who.int/classifications/en/.
TOWARDS A SEMANTIC SYSTEM FOR MANAGING CLINICAL PROCESSES
187
