
SEMANTIC ANNOTATIONS AND RETRIEVAL OF
PHARMACOBOTANICAL DATA
Ana Claudia de Macêdo Vieira
Faculdade de Farmácia, Federal University of Rio de Janeiro
Centro de Ciências da Saúde, Rio de Janeiro, Brazil
Sérgio Manuel Serra da Cruz
Estácio de Sá University – Campus WestShopping, Rio de Janeiro, Brazil
Núcleo de Computação Eletrônica, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil
Keywords: Pharmacobotany, Plant Ontology, Learning Management System.
Abstract: Images repositories would become a costly and meaningless data pool without descriptive metadata. This
paper addresses the problem capturing knowledge necessary to register and retrieve pharmacobatonical data
using semantic descriptions. We propose a Semantic Web-based Learning Management System based on
Web services to provide a semantic reasoning layer between students’ queries and stored data, our approach
enables students to handle shared data and get semantically rich results through the use of web-enabled
semantic database queries.
1 INTRODUCTION
In recent years, there has been awareness about
environmental issues and the use natural healthier
products. Scientists and governments need
informatics to support their efforts in shaping public
policies and managing natural resources. The use of
natural products derived from medicinal plants is
increasing and needs more pharmacobotanical
investigations. They demand integration of vast
amounts of information from different sources,
ranging from environmental observation to chemical
and anatomical analysis. Therefore, the study of
medicinal plants and its natural products also needs
to handle semantic data heterogeneity (Sheth, 1999).
Pharmacobotany involves morphological,
anatomical and chemical studies and they are
dependent on complete and accurate documentation
of experiment processes. Before computerization of
scientific equipments, paper notebooks were the
primary scientific record. But, with the advent of
mobile computing, satellite images and
environmental sensors the complexity of this kind of
investigations scaled and the overall numbers of
experiments performed have increased, stretching
traditional manual annotation methods to beyond
their limits. As these trends continue, and as
experiments and research teams themselves become
more distributed and cross-disciplinary, the whole
research process must become self-documenting.
Richer, more detailed, more searchable annotations
are required. Thus, metadata generated by distinct
tools used within an environmental project will have
to be integrated to provide a complete picture of the
scientific research being performed.
The semantic aspects of information integration
of data are drawing attention from research
community, and ontologies are a valuable artefact
for databases integration (Gruber et al., 1993). They
capture the semantics of information and can be
used to retrieve, annotate or store the related
metadata. Although multiple engineering artefacts
exist in the domain of Biology, do not exist to the
extent as, for instance, in teaching pharmacobotany
(Keet, 2005).
At this time, plants images, maps, textual and
multimedia files are essential part of the
pharmacobotanical scientific records. The
significance of images for identifying (plants, drugs
and anatomic details) cannot be underestimated.
333
de Macêdo Vieira A. and Serra da Cruz S. (2009).
SEMANTIC ANNOTATIONS AND RETRIEVAL OF PHARMACOBOTANICAL DATA.
In Proceedings of the First International Conference on Computer Supported Education, pages 332-337
DOI: 10.5220/0002020103320337
Copyright
c
SciTePress

Images are semantic instruments for capturing
aspects of the real world, and form a vital part of the
botanical record for which words are no substitute.
Yet, an appropriate set of images can even be used
to help to identify new natural products originated
from medicinal plants which can be further used to
either develop new formulations, reducing the
dependence from pharmaceutical laboratories or
healing the so called tropical neglected diseases
(Hotez, 2008).
In this paper, we took advantage of lessons
learned from developing ontologies in other biology
fields, most notably in Molecular Biology. To
introduce some amount of intelligence and
adaptively in this field of knowledge, we developed
a Semantic Web-based Learning Management
System (SWLMS) to enhance teaching and learning
activities. We present a SWLMS named SIM that
provides a user-friendly semantic reasoning layer
between the users and the data. It enables: teaching,
learning, collaborating with colleagues and
executing other educational activities by managing
such metadata. Section 2 describes the importance of
Pharmacobotany. Section 3 describes the semantic
approach used on the architecture. In Section 4
discusses the main characteristics of extending Plant
Ontology. Section 5 we describe a web-based
architecture used by Biology and Pharmacy
undergraduate and graduate students at Federal
University of Rio de Janeiro. Section 6 concludes
the work.
2 TEACHING
PHARMACOBOTANY
Pharmacobotany is one of very comprehensive and
complex field of human activity; it shows direct
relation with the natural living world, especially
within the plants of a given ecosystem. It strives not
only to learn about and to utilize the diversity of the
plants as widely as possible, but it also has an
interest in its preservation (Opletal, 1994).
Teaching pharmacobotany in academic
pharmacy institutions has been given new relevance,
as a result of the explosive growth in the use of
herbal remedies in modern pharmacy practice. In
turn, pharmacobotany research areas are continuing
to expand, and now include aspects of cell and
molecular biology in relation to natural products,
ethnobotany and phytotherapy, in addition to the
more traditional analytical method development,
phytochemistry and morphological and anatomical
systematization of medicinal plants.
Unfortunately, some of these fields of knowledge
still apply traditional teaching practices that consist
mostly of face-to-face lectures given by teachers;
use of scientific equipments and chemicals and, little
use of educational systems. Besides, researchers
have to use lots of images and make lots of manual
unstructured annotations about location and shape of
botanical material in environment. After that,
preparing samples, slicing it and taking another set
of images of the different parts of plant´s anatomy.
Thus, experiments execution is costly in terms of
time and materials; its results, images and
annotations are hardly ever shared.
To overcome these lacks, a SWLMS seems to be
a feasible way to engage researchers to seek new
natural products; to enhance interaction process
between learners and teachers, and to foster
collaboration in scientific community (Weitl et al.,
2002). A pharmacobotanical SWLMS can: (i) aid
teachers to plan, deliver, and manage learning
events; (ii) offer better conditions for composing and
reusing learning materials for different purposes;
(iii) enhance student’s online collaboration and
sharing annotations at lower costs.
To face the challenges of teaching such
traditional discipline, we propose a architecture
based on Semantic Web services (Berners-Lee,
2001) (Hyvonen et al., 2003) that provides a
semantic reasoning layer between users and stored
data, improving Web-based education, and
providing more independence, and intelligence from
traditional classes.
3 LEARNING MANAGEMENT
SYSTEMS AND
PHARMACOBOTANICAL
ANNOTATIONS
Learning Management Systems (LMS) have become
a broadly accepted approach to e-Learning in
universities to give support for virtual activities in
the teaching and learning processes (Devedžić,
2003). Even so, the pharmacobotanical educational
characteristics (as described in Section 2) are not yet
satisfied. There is little interoperability between
LMS; they failed to handle unstructured and
widespread heterogeneous data. Managing
annotations for botanical images has been of vital
CSEDU 2009 - International Conference on Computer Supported Education
334

importance because the value of images depends on
how easily they can be located, searched for
relevance, and retrieved. Images are usually not self-
describing.
The problem of searching large image
repositories according to their content, has been the
subject of a significant amount of research in the last
decade (Carneiro et al, 2007), some approaches are
commonly used to retrieve and annotate biological
images, such as: keywords, controlled vocabularies,
classifications and free text descriptions.
Unfortunately, they present open issues, such as the
absence to provide relations between the terms and
inheritance, which provides a controlled means to
widen or constrain a query against the repository,
they are often fraught with errors due to factors such
as annotator familiarity with the domain, amount of
training, personal motivation and complex schemas.
Thus, in order to avoid these issues, we decide to use
the ontological approach, we propose a simple
ontological model of the concepts involved in
Botany.
4 EXTENDING PLANT
ONTOLOGY
As far as we are concerned, there are no ontologies
that describe pharmacobotanical data. Thus, in order
to fulfil this gap and to support knowledge sharing
and reuse without losing interoperability, we have
extended Plant Ontology (PO).
PO is an ontology adopted on Plant Biology
scientific communities, it has been developed and
maintained with the goal to facilitate and
accommodate functional (genome and proteome)
annotation efforts in plant databases (Avraham et al,
2008). PO is not an extensive collection of botanical
terms, but rather a complex hierarchical structure in
which botanical concepts are described by their
meaning and by relationship to each other. The main
purpose of these concepts is to facilitate cross
database querying and to foster consistent use of
these vocabularies in the annotation of tissue and/or
growth stage specific expression of genes, proteins
and phenotypes. Educational aspect of the plant
ontology is to some extent limited; this is imposed
by the structure of the ontology itself and the
limitations of the current software. Thus, in order to
enrich the PO ontology concepts and augment
queries capabilities on a SWLMS, we have extended
Plant Ontology to encompass Linnaeus taxonomy
(Animal Diversity, 2008), (Legendre, Legendre,
1998). We add common pharmacobotanical
concepts describing morphological and anatomical
structures, ethobotnical and phytotherapical features
exclusive to medicinal plants. Besides, the
unambiguous classification of species represent the
foundation of scientific any Botanical research. It is
especially significant with respect to helping
scientists to understand the evolutionary process; it
identifies the fundamental divisions of life and its
progression from the simple to the complex
structures.
To expand PO we applied Kauppinen et al.
approach (2008) where association rule mining
techniques were applied to find and rank interesting
relationships based on existing pharmacobotanical
annotations, taxonomy and PO ontology. Briefly, to
find the concepts that occur often in annotations, we
therefore apply a method that consists of three
phases. First one creates the candidate relationships.
On the second phase we prune out all those
association rules that already exist in PO ontology.
On the third phase, a transitive closure is inferred for
the ontology. Hence, these phases ensure that
concepts which already have a close relationship in
ontology will not get associated again. PO extended
ontology was implemented as OWL file into
Protégé-2000 and represented as a RDF Schema.
5 SEMANTIC IMAGE
MANAGEMENT
ARCHITECTURE
The major goal of SIM is to support
pharmacobotany teaching. In this scope, ontologies
can be associated with reasoning mechanisms and
rules to enforce it. SIM was designed with some
educational goals in mind: preserve and share
knowledge about medicinal plants; support students
and teachers in managing research activities in
teaching and learning experimental
pharmacobotany; enable the adaptation to individual
learner characteristics, since no two students have
the same learning pre-requisites, skills, aptitudes or
motivations.
Plants images were created as part of
pharmacobotanical ongoing research and teaching
efforts by Ana Vieira and her team, who have also
provided us with user requirements, for instance:
browse images by species name, anatomical details,
or PO concept, preferably with images presented as
SEMANTIC ANNOTATIONS AND RETRIEVAL OF PHARMACOBOTANICAL DATA
335

thumbnails; search for all images of a given
medicinal plant or taken at a given research site;
browse images from a particular natural product.
Each image is described by a domain-specific
metadata.
5.1 SIM Goals
SIM is one of the first of what might be called
pharmacobotanical SWLMS. It combines the ideas
behind the Semantic Web and Web Services. SIM is
a Java open-source web enabled architecture, it is a
distributed architecture based on a set of semantic
Web services (Wroe et al., 2003) which aids
teachers and students to store/share educational
materials like: biological images and
pharmacobotanical annotations. SIM allow users to
retrieve images metadata and annotation through
semantic queries; browse the ontology to restore
semantically related images; recover images
according ontology’s concepts that describe plant´s
anatomical details or characteristics from its
geographical localization. All images and plant
specimen are georeferenciated (they were associated
with longitude and latitude coordinates), since
researchers and students can further return to the site
to make complimentary investigations, to take new
images or to collect new specimens. Optionally,
coordinates can be used to recover satellites images
provided by a third party service provider, like
GoogleEarth (2008) and TerraServer (2008) at very
low costs. Such approach provides researchers users
with a comprehensive aerial view of an
environment. Satellite and aerial images can be used
to manage lands and map environments. It also can
be used to monitor or analyze areas that would be
prone to having damaged during forest fires or
floods. This imagery is a unique opportunity to
expand a student’s understanding of the environment
around the medicinal plants.
5.2 SIM Architecture
In this section we introduce the SIM Conceptual
Architecture that has been designed to enable
semantics-driven Web Service applications. Figure 1
depicts the overall SIM conceptual architecture.
The architecture we distinguish three main
layers: (i) SIMWeb, it allows querying and browsing
semantics-based Web services. It’s also used by
teachers for administrating and managing the overall
system and students to retrieve data using built-in
SPARQL queries; (ii) SIMComponents, provides the
required functionality for realizing Semantic Web
enabled Web services. This layer comprises Tomcat-
Apache Server which acts as the Web services
Server; (iii) SIMStorage and external components
allow data’s persistence, mainly relying on the use
of the ontology.
Figure 1: SIM Architecture.
SIM architecture not only stores the images and
its annotations, it also registers the geographic
coordinates (latitude, longitude) of the moment of
which the plant was collect, abundance, habitat,
flowering and fruiting periods and collector’s data.
So, students are able to retrieve, at a single search all
available medicinal plants description, its
geographical position and also
GoogleEarth/TerraServer satellites point of view.
They can investigate detailed anatomical images and
annotation made at the laboratory, such approach
augments correlation between environmental and
microscopical observation of the drugs and plant
anatomy (figure 2).
Figure 2: Overview of interaction between SIM and
GoogleEarth.
Figure 3 shows a screenshot of the annotation
query interface. SIM semantic browsing has an
interesting feature; it assists users in filtering
information by selecting or combining categories,
for instance, if a student queries images and
annotations about given medicinal plant or pollinator
CSEDU 2009 - International Conference on Computer Supported Education
336
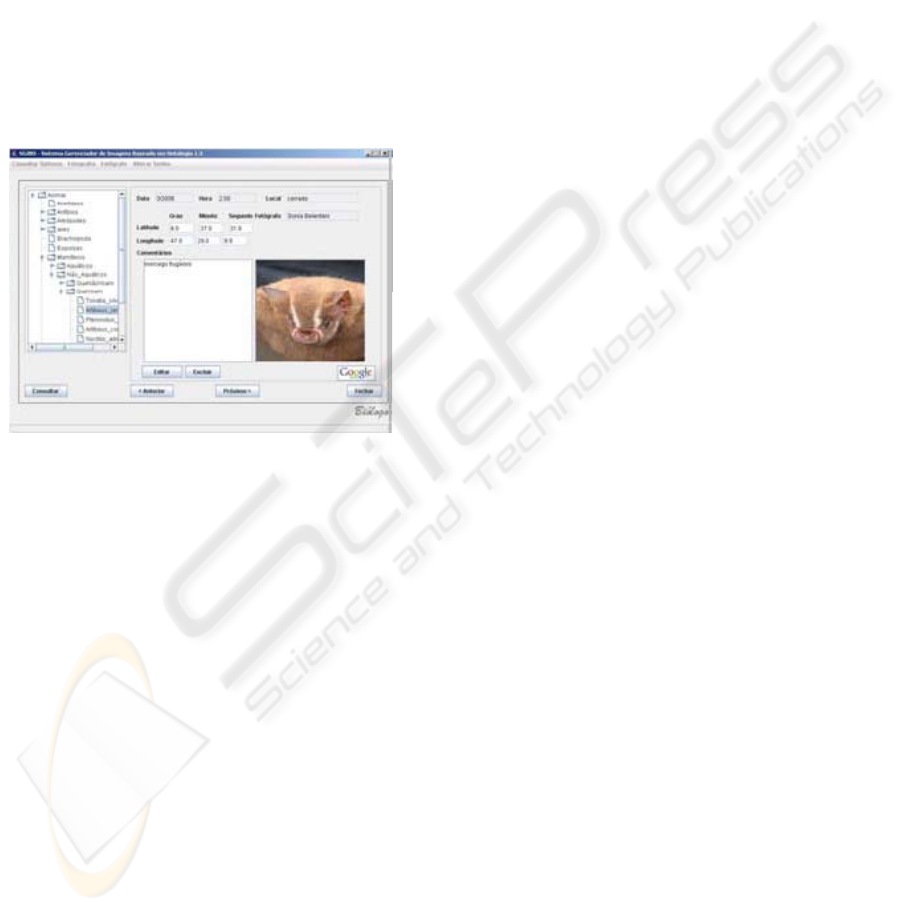
agent, an annotation tree (presented as directed
acyclic graphic) is automatically built, showing all
annotations about the plant.
One major advantage of SWLMS over a
traditional system is related with the ability to
perform concept-based searches. This type of
interaction enables students to query and manipulate
information in an intuitive manner without having to
construct logically sophisticated queries, which
requires specialised knowledge about query
languages and the underlying data model. Besides,
SIM Architecture allows students to search for
specific concepts; for example, a search for “active
principia” or “drug” will give same result even
though these are two different lexicalizations of the
same concept.
Figure 3: Annotation query interface.
6 CONCLUSIONS AND FUTURE
WORK
This paper gives some indication on how a semantic
data retrieval tool might work. Semantic annotation
allows researchers and students to make use of
concept search instead of keyword search. It paves
also the way for more advanced search strategies.
Building ontology’s for large domains, such as
Botany is a costly affair. Thus, we took advantage of
experience in modelling practices on other domains
and extended Plant Ontology to encompass
pharmacobotanical annotations. SIM Architecture
uses such extension, allowing students do make and
share annotations and retrieve semantically related
images from medicinal plants.
The contributions of this paper are threefold: (i)
it helps students to reduce the number of scientific
experiments and consequently the manipulation and
waste of hazardous and expensive chemicals once
they can collaborate by sharing and retrieve
knowledge about plants data, images or annotations
about experiments previously realized; (ii) it aids
teachers to plan, deliver, and manage learning events
that occurs outside traditional classroom, they were
also allowed them to manage students, keeping track
of their progress and performance across all types of
training activities; and (iii) it shows the feasibility of
building a Semantic Web accessible image
repository, SIM demonstrates that although existing
Semantic Web browsers do provide more flexible
user interfaces, they still have limitations in
supporting a real-world scientific usage. Some of
the missing functionalities are likely to be required
in different application contexts, such as supporting
combinations of ontological concepts; while others
are required by the challenges of presenting image
data and its metadata collected in a collaborative
way by different users.
In a near future we are going to start both
qualitative and quantitative analysis to evaluate SIM.
But, at this time we have observed improvement in
the student’s satisfaction such as through the use the
architecture we noticed an increasing ability to
integrate and share diverse sources of data. Finally,
we also noticed that students were able to perform
complex queries over the annotations.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Pedro Vieira
Cruz for his valuable collaboration and helpful
feedback. The authors gratefully acknowledge
Estácio de Sá University for the partial financial
support to this work.
REFERENCES
Animal Diversity Web. Available at:
<http://animaldiversity.ummz.umich.edu>. Last
access: 26/05/2008.
Avraham et al. S. The Plant Ontology Database: a
community resource for plant structure and
developmental stages controlled vocabulary and
annotations Nucleic Acids Research, Vol. 36.2008,
pp. 123-147.
Berners-Lee, T., Hendler, J., Lassila, O. The Semantic
Web, Scientific American, Vol. 284, No. 5, 2001, pp
34-43.
Carneiro, G., Chan, B., Moreno, P. J., Vasconcelos, N.
Supervised Learning of Semantic Classes for Image
Annotation and Retrieval, ieee transactions on pattern
SEMANTIC ANNOTATIONS AND RETRIEVAL OF PHARMACOBOTANICAL DATA
337

analysis and machine intelligence, vol. 29, no. 3, pp.
394 -410. 2007.
Devedžić, V. Key Issues in Next-Generation Web-Based
Education, IEEE Transactions on Systems, Man, and
Cybernetics – Part C: Applications and Reviews, Vol.
33, No. 3, 2003, pp. 339-349.
Google Earth. Available at <http://earth.google.com/>.
Last access: 26/07/2008.
Gruber, T R., Guarino, N., Poli, R. Toward Principles for
the Design of Ontologies Used for Knowledge
Sharing. In Formal Ontology in Conceptual Analysis
and Knowledge Representation. Kluwer Academic
Publishers, in press. Substantial revision of paper
presented at the International Workshop on Formal
Ontology, Padova, Italy, March, 1993.
Hotez, P. T. The Giant Anteater in the Room: Brazil's
Neglected Tropical Diseases Problem, PLoS Negl
Trop Dis. Vol. 2, no. 1, Jan., 2008.
Hyvonen E., Styrman A.; Saarela S. Ontology-Based
Image Retrieval. Avalilable at:
<http://www.seco.tkk.fi/publications/2002/hyvonen-
styrman-saarela-ontology-based-image-retrieval-
2003.pdf>.
Kauppinen, T., Kuittinen, H., Seppälä, K., Tuominen, J.,
Hyvönen, E. Extending an Ontology by Analyzing
Annotation Co-occurrences in a Semantic Cultural
Heritage Portal. In: ASWC 2008 Workshop on
Collective Intelligence.
Keet, C. M., Factors affecting ontology development in
ecology 2nd International Workshop on Data
Integration in the Life Sciences (DILS 2005), San
Diego. USA. 2005.
Legendre, P., Legendre L., Numerical ecology. Elsevier
Science. 2nd edition BV, Amsterdam.1998.
Opletal, L. The basis and goals of the pharmacy
profession--pharmacobotany and its contribution to
the development of drugs Ceska Slov Farm. Vol. 43,
no. 6, 1994 Nov, pp. 271-274.
Sheth, A., Changing Focus on Interoperability in
Information Systems: from System, Syntax, structure to
Semantics. In: Interoperating Geographic Information
Systems, C. Kottman, Ed. Norwell, MA: Kluwer
Academic, pp. 5-29. 1999.
Weitl, F., Süß, C., Kammerl, R., Freitag, B., Presenting
Complex e-Learning Content on the Web: A
Didactical Reference Model, In Proceedings of World
Conference on E-Learning in Corporate, Government,
Healthcare, & Higher Education, Montreal, Canada,
2002, pp. 1018-1025.
Wroe, C., Stevens, R., Goble, C., Roberts, A., Greenwood,
M.: A Suite of DAML+OIL Ontologies to Describe
Bioinformatics Web Services and Data. The
International Journal of Cooperative Information
Systems 12 pp. 597–624. 2003.
Terra Server. Available at <http://www.terraserver.com/>
Last access: 24/08/2008.
CSEDU 2009 - International Conference on Computer Supported Education
338
