
DISCOVERING RELATIONSHIP ASSOCIATIONS
IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE
Weisen Guo and Steven B. Kraines
Science Integration Program (Human), Department of Frontier Sciences and Science Integration
Division of Project Coordination, The University of Tokyo, 5-1-5 Kashiwa-no-ha, Kashiwa, Chiba, 277-8568, Japan
Keywords: Relationship Associations, Association Rules, Semantic Relationships, Semantic Matching, Semantic Web,
Ontology, Logical Inference, Life Sciences, Literature-based Knowledge Discovery.
Abstract: Over one million papers are published annually in life sciences. Bioinformatics and knowledge discovery
fields aim to help researchers conduct scientific discovery using the existing published knowledge. Existing
literature-based discovery methods and tools mainly use text-mining techniques to extract non-specified
relationships between two concepts. We present an approach that uses semantic web techniques to measure
the relevance between two relationships with specified types that involve a particular entity. We consider
two highly relevant relationships as a relationship association. Relationship associations could help
researchers generate scientific hypotheses or create computer-interpretable semantic descriptors for their
papers. The relationship association extraction process is described and the results of experiments for
extracting relationship associations from 392 semantic graphs representing MEDLINE papers are presented.
1 INTRODUCTION
The field of life sciences is one of the fastest
growing academic disciplines (Marrs and Novak,
2004). More than one million papers are published
each year in a wide range of biology and medicine
journals (King and Roberts, 1986). Recent progress
in genomics and proteomics has generated large
volumes of data on expression, function, and
interactions of gene products. As a result, there is an
overwhelming amount of experimental data and
published scientific information, much of which is
available online. Researchers in the bioinformatics
and knowledge discovery fields have been studying
how to use the existing literature to discover novel
knowledge or generate novel hypotheses.
Scientific discovery is a type of human
intellectual activity. Based on observations and
theory, researchers define hypotheses that they test
experimentally. However, due to the explosive
growth of the literature, individual scientists cannot
study all of the experimental data and scientific
information that is available.
Computational methods have been used to help
scientists generate hypotheses (Langley, 2000;
Racunas et al., 2004). For example, several attempts
have been reported to develop informatics tools that
replicate Swanson’s discovery in 1986 that fish oil
may benefit patients with Raynaud’s disease solely
from studying the literature (Swanson and
Smalheiser, 1997; Weeber et al., 2005; Hristovski et
al., 2005; Srinivasan, 2004). The possibility of
linking different scientific disciplines through
intermediate, or shared, interests has commonly
been described as Swanson’s ABC model. Most of
these literature-based discovery methods employ
text-mining techniques to find relationships of
unspecified type between two domain-related
concepts that are implied by the literature.
In this paper, we present a technique for
literature-based discovery of hypotheses based on
measuring the assocation between two relationships
of specified type that involve a particular entity or
concept. We call this a relationship association. A
relationship association is a special kind of
association rule that states “if concept A has
relationship R1 with concept B, then it is likely that
concept A has relationship R2 with concept C.”
Most scientific papers describe relationships
between concepts from the study domain, which
have been identified through research. A relationship
is essentially a semantic statement that predicates the
way in which one concept modifies the other
semantically. Our goal is to discover interesting
association rules between these relationships.
10
Guo W. and Kraines S. (2009).
DISCOVERING RELATIONSHIP ASSOCIATIONS IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE.
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval, pages 10-17
DOI: 10.5220/0002285300100017
Copyright
c
SciTePress
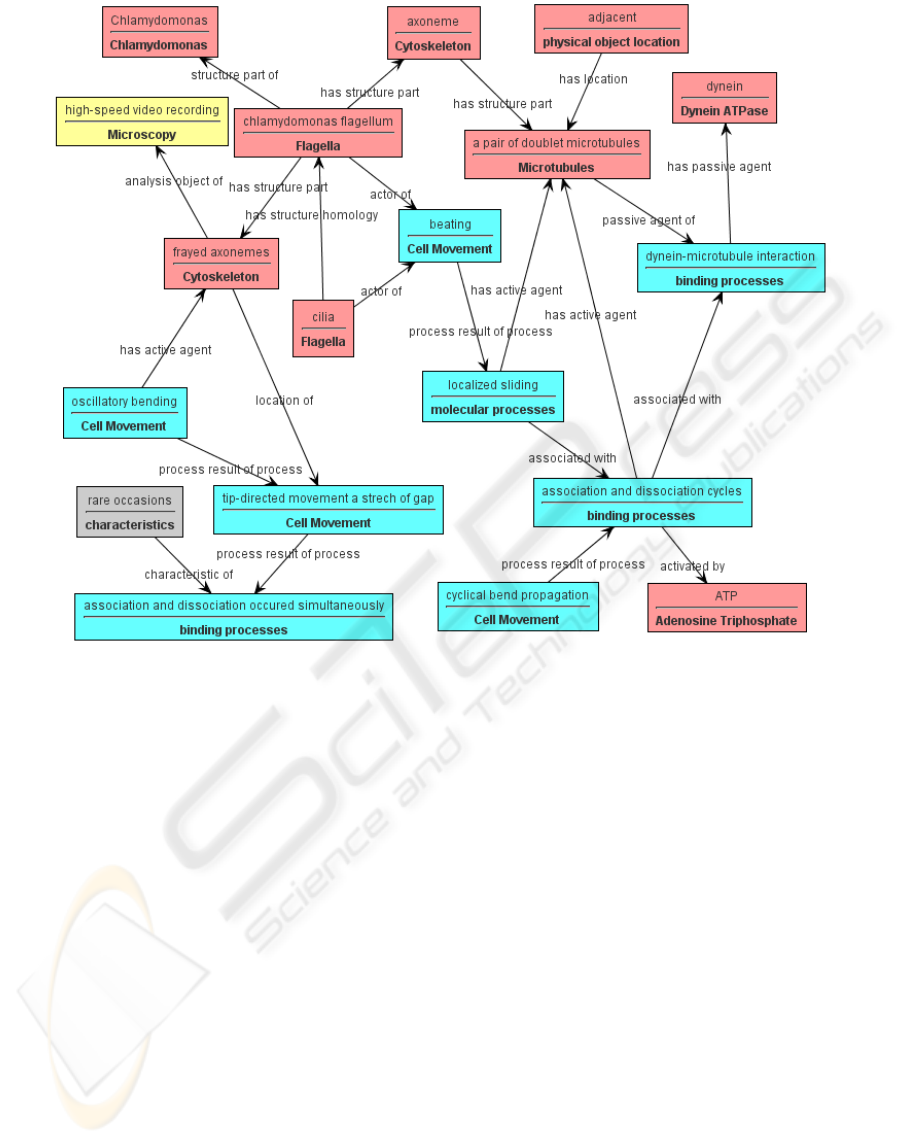
Figure 1: The Semantic graph of a paper from MEDLINE. Boxes show instances of classes from the domain ontology. The
colour of the box indicates the subsuming major upper class: blue instances are processes, red instances are physical entities,
yellow instances are investigative techniques, and gray instances are attributes. The text above the line in a box is the
instance label. The text in bold type below the line in a box is the class name of that instance. Arrows show properties
expressing the asserted relationships between instances.
Text mining techniques cannot extract
relationships between concepts with semantics that
are sufficiently precise for this kind of analysis
(Kraines 2009). We use semantic web techniques
and ontologies to define semantic relationships
described in a scientific paper as follows. First, we
create a descriptor for each paper in the form of a
semantic graph. The nodes in a semantic graph
consist of instances of particular concepts defined in
the ontology that represent entities described in the
paper. The edges in a semantic graph are the specific
relationships that the paper describes between those
entities (an example is shown in figure 1). For
example, “a Flagellum called chlamydomonas
flagellum has as a structure part a Cytoskeleton
called axoneme” is a relationship forming one arc in
the semantic graph shown in figure 1. Then, all pairs
of relationships from the semantic graphs that share
a common entity, e.g. all chains with three nodes and
two arcs, are candidates for relationship associations.
We envisage two primary usages of relationship
associations. One is helping biological scientists to
generate novel hypotheses. For example, the
relationship association that “if some kind of cellular
structure is part of some kind of flagellum, then it is
likely that the cellular structure binds to a specific
biological entity” might inspire a biologist studying
a particular kind of cellular structure, such as a
microtubule, that is part of a flagellum to generate
the hypothesis that the cellular structure binds to a
particular biological entity in the studied cell.
Relationship associations could also help users to
create computer-interpretable descriptors of their
papers in some knowledge sharing system, such as
EKOSS (Kraines et al., 2006). For example, when
the user creates a relationship describing how one
instance is modified by another, and this relationship
appears in one part of a relationship association, then
the system could automatically suggest a new
relationship and target instance to add to the instance
DISCOVERING RELATIONSHIP ASSOCIATIONS IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE
11

based on the other part of the association.
Our approach is based on two assumptions. First,
because relationship associations describe
associations of relationships between classes of
entities, we assume that similar entities have similar
relationships. Second, because we use semantic
graphs from a small part of the scientific literature to
extract the relationship associations, we assume that
if one relationship association appears in the sample
data with a high probability, then it will also appear
in the whole literature with a similar probability.
This paper is organized as follows. In Section 2,
we describe our work that forms the background for
this paper. In Section 3, we present our approach to
extract the relationship associations. In Section 4, we
describe experiments using 392 semantic graphs for
papers from MEDLINE to obtain relationship
associations. The presentation and experimental
application of the algorithm for extracting
relationship associations are the main contributions
of this paper. In Section 5, we discuss related work.
2 PRELIMINARY WORK
Many applications of semantic web technologies in
the life sciences have appeared recently, including
several large ontologies for annotating scientific
abstracts, such as the Open Biomedical Ontologies
(OBO) and the Unified Medical Language System
(UMLS) Semantic Network. In order to describe a
paper from MEDLINE as a semantic graph, we
developed the UoT ontology based on a subset of the
Medical Subject Headings (MeSH) vocabulary.
EKOSS (Expert Knowledge Ontology-based
Semantic Search) (Kraines et al., 2006) is a web-
based knowledge-sharing system that enables users
to create semantic graphs describing their knowledge
resources, such as scientific papers, using ontologies.
Figure 1 shows a semantic graph created to describe
a paper from MEDLINE (Aoyama and Kamiya,
2005). The semantic graph contains 19 instances of
classes from the UoT ontology together with 23
relationships between the instances.
In preliminary work, we have used EKOSS to
create these kinds of semantic graphs for 392 papers
selected from MEDLINE (unpublished material).
3 RELATIONSHIP ASSOCIATION
EXTRACTION
There are three main aspects to extracting
relationship associations: the data structure, the
method for determining if a relationship association
appears in a particular semantic graph, and the
algorithm for extracting the relationship associations
from a set of semantic graphs.
3.1 Semantic Graphs
The data structure determines the extracting
algorithm. Our approach uses an ontology to
represent papers semantically and unambiguously.
We use one semantic graph to represent one
MEDLINE paper. The nodes of a semantic graph are
instances of ontology classes, and the edges are
relationships between the instances that are specified
by properties also defined in the ontology. Each
instance can have a descriptive text label. Semantic
graphs, such as the one shown in figure 1, act to
structure the knowledge contained in the MEDLINE
papers for extracting relationship associations.
3.2 Semantic Matching
Matching semantic graphs is different from text
matching, such as calculating the similarity of two
strings (Cohen et al., 2003). Semantic matching
techniques compare two data structures at a semantic
level, often by using some logic inference methods.
We use a description logics reasoner software,
RacerPro (www.racer-systems.com), to evaluate the
match between a search semantic graph and a target
semantic graph through a combination of logic and
rule-based inference. First, we add the target graph
to the reasoner’s knowledge base together with the
ontology used to create the graph. Then, we convert
the search graph into a set of semantic queries by
creating sub graphs of the search graph that contain a
specified number of properties and instances. In
most cases, this is one property and two instances, i.e.
a semantic triple. Queries are created by replacing
the instances in the sub graphs with class variables.
Rules for replacing instance classes with super
classes and properties with super properties can be
applied to increase matching recall. Finally, we ask
the reasoner how many of the queries match the
target graph, where a query matches if instances in
the target graph binding to each of the class variables
in the query subject to the specified relationship(s)
can be found. The fraction of matching queries gives
the semantic similarity between the two graphs. A
simple example is shown in Figure 2. Details are
given in (Guo and Kraines, 2008).
3.3 Extraction Process
The process of extracting relationship associations
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
12
takes the set of semantic graphs as the input. The
output is a set of linked pairs of semantic
relationships, where each relationship is defined as a
triple consisting of a subject or “domain” class, an
object or “range” class, and a directed property
specifying the relationship between the two classes.
A linked pair of semantic relationships is a pair of
semantic relationships that share one class in
common. We refer to these linked pairs of semantic
relationships as relationship associations.
3.3.1 Generating Triple Queries
A semantic triple – consisting of a domain instance,
a range instance, and a property between them – is
the minimum unit of a semantic graph. One semantic
graph contains several semantic triples. The
definitions are formalized as follows:
Graph = {Triple*}
Triple = {domain,property,range}
First, for each triple in a semantic graph, we
create one triple query, defined as follows:
TripleQuery = { domain class variable,
property, range class variable}
In a triple query the instances of the triple are
converted to variables with the same classes. Thus, a
triple query converts the asserted relationship
between two specific entities made by the triple into
a generalized relationship between ontology classes.
There may be some duplicate triple queries
generated from the set of semantic graphs. However,
because we only want to link two triple queries
whose triples both appear in the same semantic
graph and share a common entity, we keep all of the
generated triple queries at this point.
3.3.2 Matching Triple Queries
We use RacerPro to infer matches between queries
and graphs via both logical and rule-based reasoning.
The logic is built into the ontology using formalisms
provided by the description logic that is supported by
the ontology specification we used (OWL-DL). The
rules are pre-defined for a particular ontology by
domain experts. Details are given in (Kraines et al.,
2006; Guo and Kraines, 2008).
If the reasoner can find a pair of instances in a
particular semantic graph meeting the class and
relationship constraints of a triple query Query1,
then we say that the triple Triple1 represented by
Query1 appears in the semantic graph. By using both
logical and rule-based reasoning, we can get
matching results that are implied at a semantic level
because the reasoner can infer relationships between
instances that are not explicitly stated in the semantic
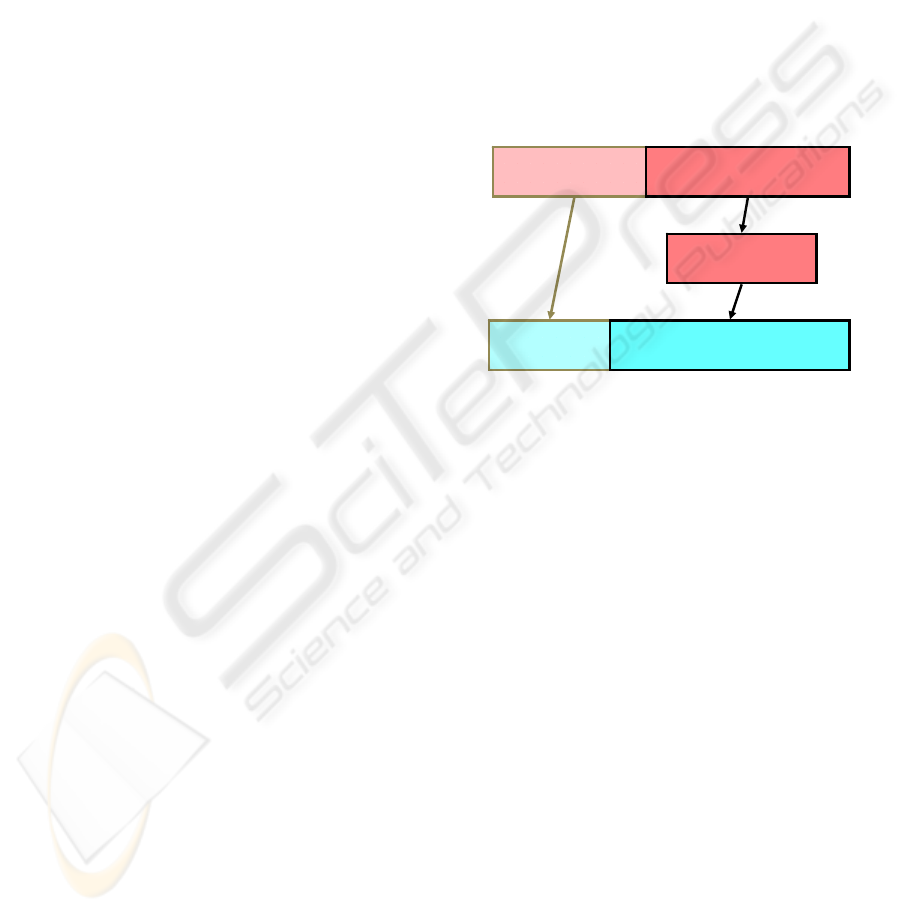
graph. For example, consider the segment of the
semantic graph in figure 1 between the instance of
Flagellum called chlamydomonas flagellum and the
instance of Cell Movement called tip-directed
movement. The triple query “find some instance of a
Flagellum that is the location of some instance of
Cell Movement” does not actually occur in the graph
because there is no property between
chlamydomonas flagellum and tip-directed
movement. However, figure 2 shows that the query
matches with the semantic graph because the
relationship is implied by the relationships specified
with the instance of Cytoskeleton called frayed
axonemes. This match is a result of the rule “If A has
structure part B and B is location of C, then A is
location of C” together with the transitivity of the
“location of” relationship.
Figure 2: An example of semantic matching. Instances are
indicated with boxes where the first line of text gives the
instance name and the second line of text gives the
instance class. Properties are shown by directed arrows
labelled with the property name. The part in outlined in
black is from the semantic graph. The part in outlined in
gray is the query.
Using the reasoner, we match all triple queries
with all semantic graphs. We then calculate the
frequency that each triple query occurs in the
semantic graphs. If a triple query only occurs in the
semantic graphs a few times, then it is not likely to
be involved in a relationship association. Therefore,
we use a user-specified threshold value to filter the
triple queries. Queries with frequencies less than the
threshold value are removed, and the rest are used to
create association queries in the next step.
3.3.3 Generating Association Queries
Now, we have a set of triple queries together with
the frequencies in which they occur in a set of
semantic graphs. In this step, we create association
queries from this set of triple queries.
For each graph, we find all pairs of triples that
share one instance and therefore comprise two
connected arcs of the semantic graph; that is, they
form a connected segment with three instances and
?
Cell Movement
?
Flagella
frayed axonemes
Cytoskeleton
chlamydomonas flagellum
Flagella
tip-directed movement
Cell Movement
has structure
part
location of
location of
DISCOVERING RELATIONSHIP ASSOCIATIONS IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE
13
two properties. If both of the corresponding triple
queries are in the set of triple queries generated in
3.3.2, then the pair of triples is a candidate for
creating an association query.
We create an association query from each triple
pair meeting the conditions above. However, to
decrease the computational load of matching them
with the set of graphs, we remove duplicate
association queries in the next step.
3.3.4 Removing Duplicate Queries
Because we use semantic matching to match an
association query with a semantic graph, two queries
with same semantic meaning will get the same
matching results. By removing association queries
with the same semantic meaning, we can reduce the
number of reasoning tasks that must be performed.
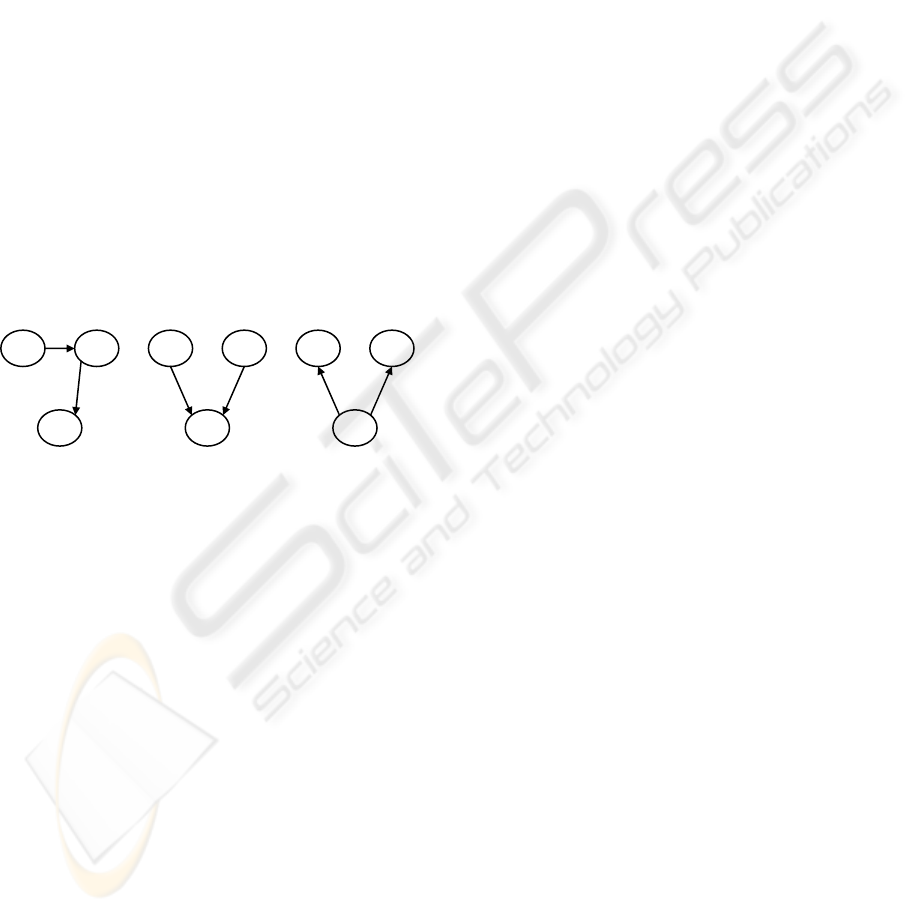
The graphs are directed, so even if two queries
have the same classes and properties, if the
directions of the properties are different, then the
queries are different. Therefore, we must consider
the three types of association queries shown in
Figure 3.
Figure 3: Three types of association queries.
We use three rules to remove duplicate
association queries. The rules are presented using a
query with type (a) as the original query Q1 (queries
with types (b) and (c) are similar):
Q1 = {v11 -> p11 -> v12; v12 -> p12 -> v13}
Each rule compares Q1 to a second query Q2 to
determine whether or not to remove Q2.
Rule 1, if Q1 and Q2 meet the following
conditions at the same time, then Q2 is removed.
z Q2 = {v21 -> p21 -> v22; v22 -> p22 -> v23}
z The class of v2i is the same as or subsumes
the class of v1i (i = 1, 2, or 3).
z The property of p2i is the same as or
subsumes the property of p1i (i = 1 or 2).
Rule 2, if Q1 and Q2 meet the following
conditions at the same time, then Q2 is removed.
z Q2 = {v21 -> p21 -> v22; v23 -> p22 -> v22}
z The class of
v2i is the same as or subsumes
the class of v1i (i = 1, 2, or 3).
z The property of p21 is the same as or
subsumes the property of p11.
z The inverse property of p22 is the same as or
subsumes the property of p12.
Rule 3, if Q1 and Q2 meet the following
conditions at the same time, then Q2 is removed.
z Q2 = {v22 -> p21 -> v21; v22 -> p22 -> v23}
z The class of v2i is the same as or subsumes
the class of v1i (i = 1, 2, or 3).
z The property of p22 is the same as or
subsumes the property of p12.
z The inverse property of p21 is the same as or
subsumes the property of p11.
As a result of this step we get a set of association
queries with unique semantics.
3.3.5 Matching Association Queries
The matching method described in Step 3.3.2 is used
to match the association queries with each of the
graphs and calculate the frequencies in which they
occur. Association queries whose frequency is less
than a given threshold are removed. The rest of the
queries are candidates for relationship associations.
3.3.6 Calculating Probabilities
From the previous steps, we get a set of association
queries meeting a specified frequency. In order to
help users find useful relationship associations, we
calculate two conditional probabilities for each
association query from the frequencies of occurrence
for the two triples that make up the association
query:
The probability that the second triple appears if
the first triple appears, prob
1-2
= P (t2 | t1).
The probability that the first triple appears if the
second triple appears, prob
2-1
= P (t1 | t2).
Generally, a high value of prob
1-2
(prob
2-1
) means
that if the first (second) triple appears in a semantic
graph, then it is likely that the second (first) triple
will also appear. If both probabilities are high, then it
is likely that the two triples will only appear at the
same time.
3.4 Relationship Associations
As a result of the extraction process described above,
we get a set of association queries together with their
probabilities of occurrence. However, this
information can be difficult for users to understand.
So we use templates and simple natural language
generation algorithms to create natural language
expressions of the relationship associations from the
association queries. The users can examine these
relationship associations to identify those that are
most reasonable and interesting. These final
relationship associations can be used to generate
v13
v12
p
11
p
12
v11
(a)
v13
v12
p
11
p
12
v11
(b)
v13
v12
p
11
p
12
v11
(c)
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
14

scientific hypotheses or to help users to create new
semantic graphs.
4 EXPERIMENTS
Using the process described above, we have
conducted experiments to obtain relationship
associations from a set of 392 MEDLINE papers. In
this section, we report the results of this experiment
As described in section 2, semantic graphs were
created for 392 papers selected from MEDLINE
using the UoT ontology that we have developed in
other work. The UoT ontology has 1,762 classes and
151 properties. We used those classes and properties
to create 392 semantic graphs. On average, each
semantic graph has 26 instances and 34 properties.
The entire set of graphs contains 10,186 instances
and 13,283 properties.
We created 13,283 triple queries from the 392
semantic graphs and then used the reasoner to
determine how many semantic graphs contain each
triple. We removed all triple queries that only
matched with one semantic graph, since that was the
graph from which the triple was obtained. As a
result, there were 8,200 triple queries available for
creating association queries.
We created 18,704 association queries based on
the 8,200 triple queries and 392 graphs. We removed
duplicates using the method from 3.3.4. We also
removed highly general queries. For example, the
property “associated with” in UoT ontology is the
top-level of the property hierarchy. Therefore, a
query containing that property does not give us any
information about the relationship type. Other highly
general “stop list” queries can be added as required.
The result is 3,483 association queries from the 392
semantic graphs.
We matched these association queries with all of
the semantic graphs using the reasoner and removed
all queries that only appeared once. This resulted in a
total of 1,215 association queries appearing in at
least two of the semantic graphs.
Next, we calculated the two probabilities prob
1-2
and prob
2-1
for each of the 1,215 association queries.
There are 629 association queries whose prob
1-2
is
greater than 0.5. There are 639 association queries
whose prob
2-1
is greater than 0.5. There are 891
association queries, for which at least one probability
(prob
1-2
or prob
2-1
) is greater than 0.5.
Finally, we converted the association queries into
natural language expressions, and we asked an
expert in life sciences to identify the most interesting
relationship associations.
Figure 4: An example of a relationship association.
One example of a relationship association that
was extracted in this experiment is shown in figure 4.
The natural language representation is: “If a
Cytoplasmic Structure is part of a Flagellum, then
the probability that there is a Physical Object that
interacts with the Cytoplasmic Structure is very
high.”
This relationship association appears in five
papers in our experiment:
“Eukaryotic flagellum is a Flagellum that has as
a part some Cellular Structure called flagellar
axoneme. The flagellar axoneme has as a part some
Microtubule called doublet microtubule that
interacts with a Dynein ATPase called dynein
arms.” (Morita and Shingyoji, 2004)
“There is a Flagellum that has as a part some
Cellular Structure called axoneme. Sliding
disintegration is a molecular process that consumes
the axonome and that is regulated by some Ion
called Ca(2+).” (Nakano et al., 2003)
“Chlamydomonas flagellum is a Flagellum that
has as a part a Cytoskeleton called axoneme. The
axoneme has as a part some Microtubule called a
pair of doublet microtubules that participates in
some binding process called dyein-microtubule
interaction. The dyein-microtubule interaction has as
a participant a Dynein ATPase called dynein.”
(Aoyama and Kamiya 2005) The semantic graph for
this paper is shown in figure 1.
“Flagellar is a Flagellum that has as a part
some Cytoplasmic Structure called
axoneme. There
is a Microtubule that is part of the axoneme. There
is a molecular process that has as an actor the
Microtubule and that is regulated by some molecule
part called dynein arm.” (Yanagisawa and Kamiya,
2004)
“There is a Flagellum that has as a part some
Cytoplasmic Structure called axoneme. Glass
substrate is a physical object that binds to the
axoneme.” (Sakakibara et al., 2004)
5 RELATED WORK
The goal of the work presented in this paper is to
discover new knowledge or hypotheses from the
Physical Objects
Cytoplasmic Structures
has structure par
t
interacts with
Flagella
DISCOVERING RELATIONSHIP ASSOCIATIONS IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE
15

literature. There are several previous attempts to
attain this goal as we mentioned in Section 1.
Swanson presented one of the first literature-
based hypotheses that fish oil may have beneficial
effects in patients with Raynaud’s disease (Swanson,
1986). His original discoveries were based on an
exhaustive reading of the literature. Swanson
described the process of his literature-based
hypotheses discovery with his ABC model: if A and
B are related, and B and C are related, he suggested
that A and C might be indirectly related.
The text analysis scripts developed from
Swanson’s initial work evolved into the Arrowsmith
system (Swanson and Smalheiser, 1997). The
Arrowsmith system considers the titles of papers
from MEDLINE. If two concepts co-occur in a title,
then they are considered to be related. Therefore, the
Arrowsmith system uses the relationships of co-
occurrence of concepts in titles to infer the implicit
relationships between two concepts.
Gordon and Lindsay developed a methodology
for replicating Swanson’s discovery based on lexical
statistics. They used different word frequency-based
statistics, including words and multiword phrases
from entire MEDLINE records in addition to title
words (Gordon and Lindsay, 1996; Lindsay and
Gordon, 1999).
Weeber and colleagues used the Unified Medical
Language System (UMLS) Metathesaurus to identify
biomedically interesting concepts in MEDLINE
titles and abstracts. They also exploited the semantic
categorisation that is included in the UMLS
framework (Weeber et al., 2003, 2005).
Hristovski and colleagues used the manually
assigned MeSH terms rather than the natural
language text from MEDLINE citations. Their tool
BITOLA computes association rules between MeSH
terms. They used association rules to measure the
relationship between MeSH term concepts in the
form X -> Y (confidence, support). They used
concept co-occurrence as an indication of a
relationship between concepts, but they did not try to
identify the kind of relationship. Therefore, although
their association rule method determines whether or
not there is implicit relationship between two
concepts, it cannot identify the specific type of
relationships that are associated. Their association
rules are between two concepts, not two
relationships (Hristovski et al., 2001, 2005).
All these existing approaches focus on extracting
non-specified relationships between two concepts in
the target domain. In contrast, our approach tries to
discover an implicit association between a pair of
relationships, each of which predicates the specific
way that one concept modifies another. We call a
pair of relationships that are found to be relevant a
relationship association. Our approach uses semantic
web techniques to enable this kind of discovery of
implied associations between relationships.
Although Hristovski et al. suggested that MeSH
terms represent more precisely what a particular
document is about than plain text, MeSH terms
cannot represent the relationships between the
entities that are described. Our approach uses
concepts and properties specified in an ontology that
logically structures a set of MeSH terms in order to
represent the relationships between entities described
in a MEDLINE paper, which we believe provides a
more precise representation of that paper.
6 CONCLUSIONS
How to help researchers make scientific discoveries
using the existing published knowledge is an
important problem in bioinformatics and knowledge
discovery fields. Recently, many literature-based
discovery methods and tools have been proposed for
solving this problem. These approaches mainly use
text-mining techniques to discover non-specified
relationships between two concepts.
We have presented an approach based on
semantic web techniques to discover the association
of pairs of specified relationships, which we call
relationship associations. These relationship
associations could help researchers generate
scientific hypotheses and also assist in the creation
of semantic graphs describing scientific documents
in a computer-interpretable way.
We first reviewed our preliminary work for
creating semantic graphs using an ontology
developed from a subset of the MeSH vocabulary.
Then, we described the process of extracting
relationship associations from those semantic graphs.
First, we generate triple queries from the semantic
graphs and calculate their frequencies of occurrence
by matching them with the set of semantic graphs
using logical and rule-based inference. Next, we
generate association queries from the triple queries
whose frequencies of occurrence are larger than a
specified threshold. We remove association queries
that specify the same semantic relationships and
match the remaining association queries with the set
of semantic graphs to get their frequencies of
occurrence. Finally, we convert the association
queries whose frequencies exceed the given
threshold to relationship associations expressed in
natural language.
We discussed the results of an experiment to
apply the approach to a set of 392 semantic graphs
based on papers from MEDLINE. The relationship
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
16

associations that were created from these semantic
graphs were examined and several interesting ones
were identified.
The relationship association extraction method
presented here can be used in other knowledge
domains. In future work, we plan to apply the
method to extract relationship associations from a set
of semantic graphs that have been created to express
failure events in the field of engineering.
ACKNOWLEDGEMENTS
The authors thank Daisuke Hoshiyama for advice on
interpretation of the experimentally extracted
relationship associations and the President’s Office
of the University of Tokyo for funding support.
REFERENCES
Aoyama, S., Kamiya, R., 2005. Cyclical Interactions
between Two Outer Doublet Microtubules in Split
Flagellar Axonemes. Biophys J., 89 (5), 3261-3268.
Cohen, W.W., Ravikumar, P., Fienberg, S.E., 2003. A
Comparison of String Distance Metrics for Name-
Matching Tasks. Proceedings of the ACM Workshop
on Data Cleaning, Record Linkage and Object
Identification, Washington DC, August 2003.
Gordon, M. D., Lindsay, R.K., 1996. Toward Discovery
Support Systems: A Replication, Re-Examination, and
Extension of Swanson’s Work on Literature-Based
Discovery of a Connection between Raynaud’s and
Fish Oil. JASIST, 47(2), 116-128.
Guo, W., Kraines, S., 2008. Explicit Scientific Knowledge
Comparison Based on Semantic Description Matching.
American Society for Information Science and
Technology 2008 Annual Meeting, Columbus, Ohio.
Hristovski, D., Stare, J., Peterlin, B., Dzeroski, S., 2001.
Supporing discovery in medicine by association rule
mining in Medline and UMLS, Medinfo, 10(Pt2),
1344-1348.
Hristovski, D., Peterlin, B., Mitchell, J.A., Humphrey,
S.M., 2005. Using literature-based discovery to
identify disease candidate genes. International Journal
of Medical Informatics, 74(2-4), 289-298.
King, T.J., Roberts, M.B.V, 1986. Biology: A Functional
Approach. Thomas Nelson and Sons. ISBN 978-
0174480358.
Kraines, S., Guo, W., Kemper, B., Nakamura, Y., 2006.
EKOSS: A Knowledge-User Centered Approach to
Knowledge Sharing, Discovery, and Integration on the
Semantic Web. The 5th International Semantic Web
Conference, LNCS 4273, 833-846.
Kraines, S., 2009. An Ontology-based System for Sharing
Expert Knowledge in Life Sciences. Journal of Web
Semantics, in review.
Langley, P., 2000. The computational support of scientific
discovery. International Journal of Human-Computer
Studies, 53, 393-410.
Lindsay, R.K., Gordon, M.D., 1999. Literature-based
discovery by lexical statistics, JASIST, 50 (7), 574-587.
Marrs, K.A., Novak, G., 2004. Just-in-Time Teaching in
Biology: Creating an Active Learner Classroom Using
the Internet. Cell Biology Education, 3, 49-61.
Morita, Y., Shingyoji, C., 2004. Effects of imposed
bending on microtubule sliding in sperm flagella.
Current Biology, 14(23), 2113-2118.
Nakano, I., Kobayashi, T., Yoshimura, M., Shingyoji, C.,
2003. Central-pair-linked regulation of microtubule
sliding by calcium in flagellar axonemes. Journal of
Cell Science, 116 (8), 1627-1636.
Racunas, S.A., Shah, N.H., Albert, I., Fedoroff, N.V., 2004.
HyBrow: a prototype system for computer-aided
hypothesis evaluation. Biofinformatics, 20 (Suppl 1),
i257-i264.
Sakakibara, H.M., Kunioka, Y., Yamada, T., Kamimura,
S., 2004. Diameter oscillation of axonemes in sea-
urchin sperm flagella. Biophys J.
, 86(1 Pt 1), 346-352.
Srinivasan, P., 2004. Text Mining: Generating Hypotheses
From MEDLINE. JASIST, 55(5), 396-413.
Swanson, D.R., 1986. Fish oil, Raynaud’s syndrome, and
undiscovered public knowledge. Perspectives in
Biology and Medicine, 30, 7-18.
Swanson, D.R., 1988. Migraine and Magnesium: Eleven
neglected connections. Perspectives in Biology and
Medicine, 31, 526-557.
Swanson, D.R., 1990. Somatomedin C and Arginine:
Implicit connections between mutually isolated
literatures. Perspectives in Biology and Medicine,
33(2), 157-179.
Swanson, D.R., Smalheiser, N.R., 1997. An interactive
system for finding complementary literatures: a
stimulus to scientific discovery. Artificial Intelligence,
91, 183-203.
Swanson, D. R., Smalheiser, N.R., Bookstein, A., 2001.
Information discovery from complementary literatures:
Categorizing viruses as potential weapons. JASIST,
52(10), 797-812.
Weeber, M., Vos, R., Klein, H., de Jong-van den Berg,
L.T.W, Aronson, A.R, Molema, G., 2003. Generating
hypotheses by discovering implicit associations in the
literature: A case report of a search for new potential
therapeutic uses for thalidomide, In J. American
Medical Informatics Association, 10(3), 252-259.
Weeber, M., Kors, J.A., Mons, B., 2005. Online tools to
support literature-based discovery in the life sciences.
Briefings in Bioinformatics, 6(3), 277-286.
Yanagisawa, H., Kamiya, R., 2004. A Tektin Homologues
Is Decreased in Chlamydomonas Mutants Lacking an
Axonemal Inner-Arm Dynein. Molecular Biology of
the Cell, 15 (5), 2105-2115.
DISCOVERING RELATIONSHIP ASSOCIATIONS IN LIFE SCIENCES USING ONTOLOGY AND INFERENCE
17
