
LINK INTEGRATOR
A Link-based Data Integration Architecture
Pedro Lopes, Joel Arrais and José Luís Oliveira
Universidade de Aveiro, DETI/IEETA, Campus Universitário de Santiago, 3810 – 193 Aveiro, Portugal
Keywords: Data integration, Link integration, Service integration, Web application, Web2.0, Lsdb.
Abstract: The evolution of the World Wide Web has created a great opportunity for data production and for the
construction of public repositories that can be accessed all over the world. However, as our ability to
generate new data grows, there is a dramatic increase in the need for its efficient integration and access to
all the dispersed data. In specific fields such as biology and biomedicine, data integration challenges are
even more complex. The amount of raw data, the possible data associations, the diversity of concepts and
data formats, and the demand for information quality assurance are just a few issues that hinder the
development of a general proposal and solid solutions. In this article we describe a lightweight information
integration architecture that is capable of unifying, in a single access point, several heterogeneous
bioinformatics data sources. The model is based on web crawling that automatically collects keywords
related with biological concepts that are previously defined in a navigation protocol. This crawling phase
allows the construction of a link-based integration mechanism that conducts users to the right source of
information, keeping the original interfaces of available information and maintaining the credits of original
data providers.
1 INTRODUCTION
World Wide Web and its associated technologies are
evolving rapidly as is the ability to develop and
deploy customized solutions for users. It is
becoming easier over time to create novel
applications with attractive interfaces and advanced
features. However, this evolution leads to several
problems. The major problem is in finding
information with certified quality: being extremely
easy to deploy new content online, the amount of
incorrect and invalid information is growing. This
process also leverages content heterogeneity. The
same concept can be stored in different media
formats or database models and accessed by
different kinds of methods. Data integration
architectures propose a single unifying framework
that can connect to several distinct data sources and
offer their content to users through web applications,
remote services or any other kind of data exchange
API.
Despite the advances in application development
and web standards, integrating information is still a
challenging task. This difficulty can be mostly seen
in life sciences. Biology and biomedicine are using
information technologies to solve their problems or
share new discoveries. The Human Genome Project
– HGP (Collins et al., 1998) – required novel
software applications that could help in solving
biological problems faster, ease biologists’ everyday
tasks, aid in the publication of relevant scientific
discoveries and promote researchers’
communication and cooperation. This exponential
necessity empowered a new field of research
denominated bioinformatics, requiring combined
efforts from biologists, statisticians, and software
engineers. The success of HGP brought about a new
wave of research projects (Adams et al., 1991,
Cotton et al., 2008) that resulted in a dramatic
increase of information available online.
With the tremendous amount of available data,
integration is the most common issue when
developing new solutions in the area of
bioinformatics. Applications like UniProt (Bairoch
et al., 2005) and Ensembl (Hubbard et al., 2002) or
warehouses like NCBI (Edgar et al., 2002, Pruitt and
Maglott, 2001) aim to integrate biological and
biomedical data that is already available. The
applied data integration strategies are suitable when
the purpose is to cover large data sources that use
standard data transfer models. However, when the
274
Lopes P., Arrais J. and Luís Oliveira J. (2009).
LINK INTEGRATOR - A Link-based Data Integration Architecture.
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval, pages 274-277
DOI: 10.5220/0002291702740277
Copyright
c
SciTePress

data is presented in pure HTML, REST web
services, CSV sheets or plain text, the integration
process can be quite complex. Along with this
problem there is the fact that when small
applications are integrated in this environment, the
content authorship is lost. This means that the
original researcher, who made a great effort to
publish his work online, will be hidden behind a
small link contained in a simple list among similar
links. The architecture described in this paper solves
both these problems. We propose an integration
model that can easily integrate information by
storing its URL and displaying it inside a centralized
application. With this, the data integration problem
is partially solved and the original application is
presented and extended to users.
The following section debates Link Integrator
organization and section 3 describe a real world
implementation scenario. Finally, section 4 contains
some final remarks about our research.
2 LINK INTEGRATOR
To deal with the presented integration issues, one
can adopt several distinct strategies for data
integration. These approaches differ mostly on the
amount and kind of data that is merged in the central
database. Different architectures will also generate a
different impact on the application performance and
efficiency.
Warehouse (Polyzotis et al., 2008, Reddy et al.,
2009, Zhu et al., 2008) solutions consist in
replicating integrated data sources in a single
physical location with a unifying data model.
Mediator-based solutions (Haas et al., 2001) rely on
a middleware layer for the creation of a proxy
between the client and the original integrated
servers. Link-based integration (Maglott et al., 2007,
Oliveira et al., 2004) strategies simply list URLs
linking the original data sources. Arrais work (Arrais
et al., 2007) presents a solid analysis on these
strategies, their main advantages and disadvantages,
resulting in an optimal hybrid solution.
In addition to these strategies, current trends
involve developments in the meta-applications area.
Mashups (Belleau et al., 2008) and workflows (Oinn
et al., 2004) have a growing popularity among data
and service integration architectures. When the goal
is to offer real-time web-based dynamic integration,
with an increased user-control, Lopes’ work (Lopes
et al., 2008) presents a valuable solution that can be
adapted to several scenarios.
We have chosen to design a solution based on
link integration scenarios. Our choice is mostly due
to the fact that the biological content available
online is dynamically changing and evolving swiftly.
2.1 Architecture
Link Integrator architecture metaphor relies on
typical three-layer architecture. The proposed
architecture divides the application structure: data
access, application logic and presentation layer.
The data access layer is completely independent
from our application. It represents the external data
sources that are accessed by originally integrated
applications. The application logic layer is crucial in
our system: externally it deals with the
communications with the integrated applications;
internally it deals with the application execution
cycle. The presentation layer is where any user in his
web browser can access a single unifying interface,
designed to be attractive and fulfilling high quality
usability patterns.
It is important to highlight that our system does
not have any kind of communication or data
exchange with the original data sources. This type of
activity would breach the initial system
requirements, transforming the integration model
from link-based to mediator-based.
2.2 Execution
Link Integrator is composed of a map, containing
navigational information that the crawler will read to
configure its processing cycle; a topic driven web
crawler capable of parsing and processing the URLs
it gathers and a relational database to store the
information gathered by the crawler.
In this system, the most important part is the web
crawler. The map contains mostly URL addresses
and regular expressions that are used by the crawler
to retrieve, parse and process web documents. The
crawling results are triplets containing an association
between: an Entity, which is the generic category
where the found URLs will fit; a Class that
represents the container where the link belongs; a
link to the Application, the URL, which is the main
result of the crawling cycle. Both the Entities and
the Classes are defined in the map; the crawler only
finishes the triplet by associating the URL with the
Class and with the Entity.
The system operation cycle – Figure 1 – is
simple. Initially, the system reads a predefined
navigation map containing Entities, Classes, URLs
and regular expressions to find other URLs. The
crawler will then parse and process the web pages
generating sets of results – URL-Class-Entity triplets
– that will be stored in containers. These containers
LINK INTEGRATOR - A Link-based Data Integration Architecture
275
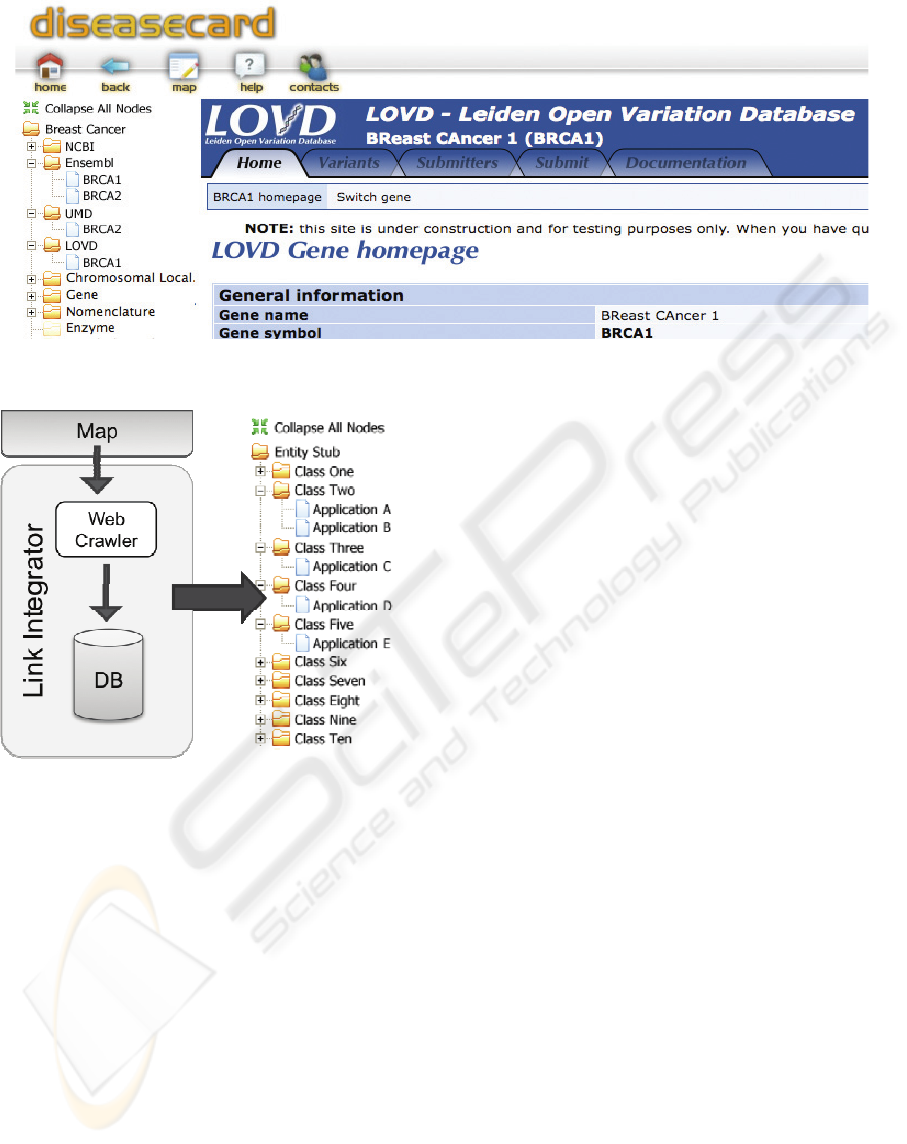
Figure 2: DiseaseCard interface with Link Integrator information.
Figure 1: Link Integrator execution workflow.
can be serialized in XML and passed to another
application or, as we do, stored in a relational
database. After this crawling and retrieving phase,
the application engine generates, from the data
stored in the relational database, a tree that is
presented to the user in the application’s main
workspace.
3 DISEASECARD
DiseaseCard is a link-based integration application
with the main purpose of connecting and presenting
disease related information spread throughout the
Internet. It was first launched in 2005 and has grown
to a mature state where up to 2000 rare diseases
stored in the Online Mendelian Inheritance in Man
(Hamosh et al., 2005) database are covered.
When DiseaseCard was created, web
applications where mostly based on static HTML
layouts to display content. Along with Internet
evolution, web application complexity has also
evolved: Web2.0 dynamic applications and web
service responses cannot be parsed by traditional
web crawlers.
Locus specific databases contain information
that is as important for researchers as the
information already gathered in DiseaseCard. These
databases store information about genetic variations
and relating this information with other genetics
marks can offer newer insights on rare disease
studies. Joining DiseaseCard paradigm with amount
of information contained in LSDBs we have the
perfect test bed for Link Integrator. Adding LSDBs
to DiseaseCard will increase the added value it
represents in the biomedical community and will
validate the goals we set when developing our
system. The new on development version of
DiseaseCard – Figure 2 – will be supported by the
Link Integrator engine that can join information
from several LSBDs applications like LOVD
(Fokkema et al., 2005) or UMD (Al and Junien,
2000). The current portal is available at
www.diseasecard.org.
4 CONCLUSIONS
Taking into account the amount of data available
after the Human Genome Project, the area of life
sciences is probably the discipline most affected by
information quantity, diversity and heterogeneity.
Recently, an area of growing importance is related to
the human variome. Variation studies often result in
KDIR 2009 - International Conference on Knowledge Discovery and Information Retrieval
276

new web applications denominated locus specific
databases and they usually contain information
about sequence variations among individuals for a
particular gene. In addition, content ownership and
its growing importance is gaining relevance. Despite
the fact that for regular end-users, access to
scientific content is easier when provided by a
centralized service, researchers who want to publish
their work are almost obliged to create their own
applications if they want to keep the authorship of
their work visible.
The described architecture and application intend
to overcome these problems with three key features
for both users and researchers. First, integration is
based on simple Internet URLs that are parsed and
processed to gather the most significant information.
This means that developers will not have to make
any changes to the application core and that we are
able to integrate any URL-accessible content.
Secondly, the original applications will be shown
inside our application. Thus, the content owners will
not be shown as a link but as part of a complete
application. Finally, external applications can be
extended inside our system: information exchanges,
text-mining and other user customization features
can be developed to enhance the original
applications.
ACKNOWLEDGEMENTS
The research leading to these results has received
funding from the European Community's Seventh
Framework Programme (FP7/2007-2013) under
grant agreement nº 200754 - the GEN2PHEN
project.
REFERENCES
Adams, M. D., Kelley, J. M., et al. (1991) Complementary
DNA sequencing: expressed sequence tags and human
genome project. Science, 252, 1651-1656.
Al, B. E. T. & Junien, C. (2000) UMD (Universal
Mutation Database): A Generic Software to Build and
Analyze Locus-Specific Databases. Human Mutation,
94.
Arrais, J., Santos, B., et al. (2007) GeneBrowser: an
approach for integration and functional classification
of genomic data. Journal of Integrative
Bioinformatics, 4.
Bairoch, A., Apweiler, R., et al. (2005) The Universal
Protein Resource (UniProt). Nucleic Acids Research,
33, 0-159.
Belleau, F., Nolin, M.-A., et al. (2008) Bio2RDF: Towards
a mashup to build bioinformatics knowledge systems.
Journal of Biomedical Informatics, 41, 706-716.
Collins, F. S., Patrinos, A., et al. (1998) New Goals for the
U.S. Human Genome Project: 1998-2003. Science,
282, 682-689.
Cotton, R. G. H., Auerbach, A. D., et al. (2008)
GENETICS: The Human Variome Project. Science,
322, 861-862.
Edgar, R., Domrachev, M. & Lash, A. E. (2002) Gene
Expression Omnibus: NCBI gene expression and
hybridization array data repository. Nucleic Acids
Research, 30, 207-210.
Fokkema, I. F., Den Dunnen, J. T. & Taschner, P. E.
(2005) LOVD: easy creation of a locus-specific
sequence variation database using an "LSDB-in-a-
box" approach. Human Mutation, 26, 63-68.
Haas, L. M., Schwarz, P. M., et al. (2001) DiscoveryLink:
A system for integrated access to life sciences data
sources. IBM Systems Journal, 40, 489-511.
Hamosh, A., Scott, A. F., et al. (2005) Online Mendelian
Inheritance in Man (OMIM), a knowledgebase of
human genes and genetic disorders. Nucleic Acids
Research, 33, 514-517.
Hubbard, T., Barker, D., et al. (2002) The Ensembl
genome database project. Nucleic Acids Research, 30,
38-41.
Lopes, P., Arrais, J. & Oliveira, J. L. (2008) Dynamic
Service Integration using Web-based Workflows.
Proceedings of the 10th Internation Conference on
Information Integration and Web Applications &
Services. Linz, Austria, Association for Computer
Machinery.
Maglott, D., Ostell, J., et al. (2007) Entrez Gene: gene-
centered information at NCBI. Nucleic Acids
Research, 35.
Oinn, T., Addis, M., et al. (2004) Taverna: a tool for the
composition and enactment of bioinformatics
workflows. Bioinformatics, 20, 3045-3054.
Oliveira, J. L., Dias, G. M. S., et al. (2004) DiseaseCard:
A Web-based Tool for the Collaborative Integration of
Genetic and Medical Information. Proceedings of the
5th International Symposium on Biological and
Medical Data Analysis, ISBMDA 2004. Barcelona,
Spain, Springer.
Polyzotis, N., Skiadopoulos, S., et al. (2008) Meshing
Streaming Updates with Persistent Data in an Active
Data Warehouse. Knowledge and Data Engineering,
IEEE Transactions on, 20, 976-991.
Pruitt, K. D. & Maglott, D. R. (2001) RefSeq and
LocusLink: NCBI gene-centered resources. Nucleic
Acids Research, 29, 137-140.
Reddy, S. S. S., Reddy, L. S. S., et al. (2009) Advanced
Techniques for Scientific Data Warehouses. Advanced
Computer Control, 2009. ICACC '09. International
Conference on.
Zhu, Y., An, L. & Liu, S. (2008) Data Updating and Query
in Real-Time Data Warehouse System. Computer
Science and Software Engineering, 2008 International
Conference on.
LINK INTEGRATOR - A Link-based Data Integration Architecture
277
