
SIABO
Semantic Information Access through Biomedical Ontologies
Troels Andreasen, Henrik Bulskov, Tine Lassen, Sine Zambach
CBIT, Roskilde University, Universitetsvej 1, Roskilde, Denmark
Per Anker Jensen, Bodil Nistrup Madsen, Hanne Erdman Thomsen
ISV, Copenhagen Business School, Dalgas Have 15, Frederiksberg, Denmark
Jørgen Fischer Nilsson, Bartlomiej Antoni Szymczak
IMM, Technical University of Denmark, Richard Petersens Plads, Kongens Lyngby, Denmark
Keywords: Domain modelling, Ontology engineering, Natural language processing, Ontological, Content-oriented text
search.
Abstract: The scientific aim of the project presented in this paper is to provide an approach to representing,
organizing, and accessing conceptual content of biomedical texts using a formal ontology. The ontology is
based on UMLS resources supplemented with domain ontologies developed in the project. The approach
introduces the notion of ‘generative ontologies’, i.e., ontologies providing increasingly specialized concepts
reflecting the phrase structure of natural language. Furthermore, we propose a novel so-called ‘ontological
semantics’ which maps noun phrases from texts and queries into nodes in the generative ontology. This
enables an advanced form of data mining of texts identifying paraphrases and concept relations and
measuring distances between key concepts in texts. Thus, the project gains its identity in its attempt to
provide a formal underpinning of conceptual similarity or relatedness of meaning.
1 INTRODUCTION
Search in texts is progressing beyond conventional
keyword search in order to make it less syntactic and
more semantically oriented. This paper presents
endeavours in the SIABO project aiming at
achieving content-based text search within the
application area of biomedicine.
Our main thesis is that a content-based search
functionality can be achieved by computerised text
analysis using ontologies enhanced with domain
models and language processing.
The remainder of this section describes the aims
of the SIABO project in general, section 2
introduces the notion of ‘generative ontology’,
section 3 presents the kind of domain modelling
carried out in the project, section 4 sets out two
approaches to concept extraction which we are
currently testing, one synthetic, and the other
pattern-based. Section 5 addresses the problems
related to querying information and knowledge, and
finally, in section 6, we present our conclusions.
1.1 The SIABO Project
The aim of the SIABO project is to provide an
approach to representing, organizing, and accessing
the conceptual content of biomedical texts using a
formal ontology.
In order to be competitive, companies need to
have access to the contents of the increasing amount
of documentation about their products, processes
and projects. Retrieval of information and
knowledge from huge, diverse resources is vital, and
only a semantics-based approach to information
management is adequate to that task.
This project presents an approach in which the
meaning content of each document is described as a
set of arbitrarily complex conceptual feature
structures facilitating detailed comparison of the
171
Andreasen T., Bulskov H., Lassen T., Zambach S., Anker Jensen P., Nistrup Madsen B., Erdman Thomsen H., Fischer Nilsson J. and Antoni Szymczak
B. (2009).
SIABO - Semantic Information Access through Biomedical Ontologies.
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development, pages 171-176
DOI: 10.5220/0002297801710176
Copyright
c
SciTePress

content of documents. The properties of an
ontology-based system lead to easier access to data
sources, locally as well as globally.
Ontologies are formal tools for structuring the
concepts of a scientific domain by means of
relationships between concepts, e.g., along the
specialization/generalization dimension. The SIABO
approach introduces the notion of generative
ontologies, i.e., infinite ontologies providing
increasingly specialized concepts. The project sets
up a novel, so-called ’ontological semantics‘, which
maps the conceptual content of phrases into points in
the generative ontology. Text chunks with identical
meaning but different linguistic forms are to be
mapped to the same node in the generative ontology.
Thus, the approach facilitates identification of
paraphrases, conceptual relationships and
measurement of distances between key concepts in
texts. The project focuses on ontological engineering
of biomedical ontologies applying lattices and
relation-algebras, and has clear affinities to
contemporary research in the Semantic Web area,
description logic as well as XML approaches.
However, it gains its distinct innovative scientific
profile by means of the above-mentioned notions.
2 GENERATIVE ONTOLOGY
A generative ontology is based on a finite ontology
with the isa concept inclusion relation (called the
‘skeleton ontology’), enriched with a set of semantic
relations providing generativity. For instance, the
skeleton ontology may specify the inclusion path:
insulin secretion isa secretion isa process isa event
A generative ontology is to be understood as a
non-finite set of concepts. We move from finite
ontologies to infinite systems of concepts, thereby
reflecting the recursive productivity of the phrase
structures in natural language. This makes it possible
to map complex linguistic structures into
correspondingly complex concepts associated with
nodes in the ontology.
Semantic relations provide feature structures
such as disease[CausedBy: lack[WithRespectTo:
insulin]], which corresponds to linguistic forms
found in a text or a query, such as diseases caused
by insulin lack, diseases induced by insulin
deficiency, insulin deficiency disease, etc.
2.1 Concept Feature Structures
Concept feature structures are recursive structures,
taking the following form:
c[r
1
:c
1
, r
2
:c
2
, …, r
n
:c
n
]
where c is a concept from the skeleton ontology, and
r
1
, r
2
, … are semantic relations, and c
1
, c
2
, … c
n
are
concept feature structures. Note that an atomic
concept is also a concept feature structure.
The attributions (feature-value pairs) [r
1
:c
1
, r
2
:c
2
, …]
consist of relations and concept arguments, and
function as conceptual restrictions on the head
concept c. This means that c[r
1
:c
1
] is always situated
below the node c in the ontology. In this way, new
paths stretch towards more specialised concepts in
the ontology. However, the generative ontology does
not admit arbitrary combinations of relations and
concepts: The relations function as case roles (cf.
Fillmore, 1968) expressing ontologically admissible
ways of combining concepts, according to so-called
‘ontological affinities’. Currently, logical affinities
are specified as triples <c’,r,c’’>. The affinities are
specified so as to rule out category mistakes. In our
context of ontologies for scientific texts within bio-
medicine, we concentrate on physical-chemical-
biological categories and disregard metaphors.
3 DOMAIN ONTOLOGIES
As a validated fragment of the generative ontology,
we construct domain ontologies supplementing and
refining already existing ontologies for the domain,
such as UMLS (Unified Medical Language System).
Validated domain ontologies are needed because
UMLS is not specific enough as regards concepts
and concept relations, and in many cases, the
existing resources are imprecise. By adopting the
principles of terminological ontologies and by
consulting domain experts, we arrive at validated
concept structures. As the basis of the domain
ontologies, a small text corpus has been used to
produce a list of term candidates, and in cooperation
with a domain expert, central concepts have been
identified. Furthermore, UMLS resources have been
consulted.
3.1 Terminological Ontologies
In this project, the domain ontologies are
terminological ontologies, i.e., their structure is
based on characteristics and subdivision criteria
Madsen et al., 2005), and we use an extended set of
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
172
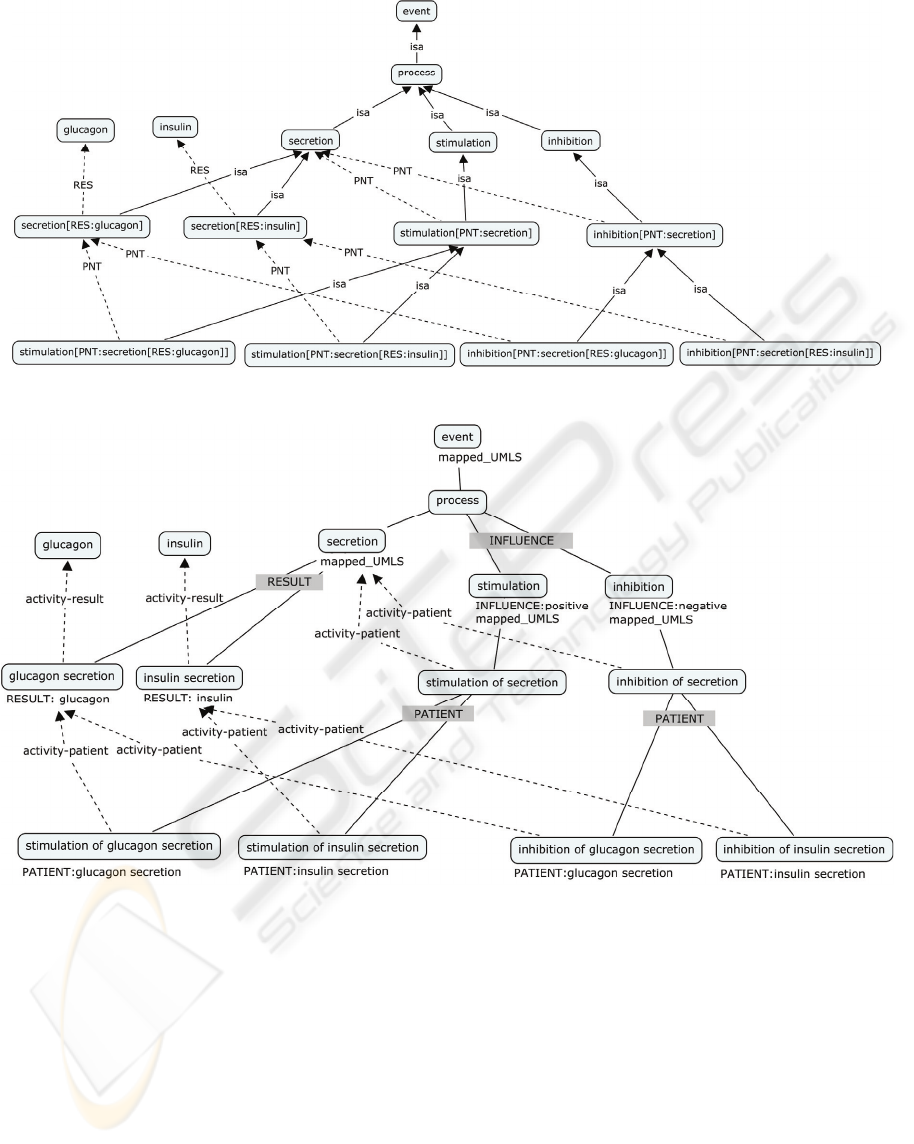
Figure 1: Extract of a generative ontology of insulin production.
Figure 2: Extract of the domain ontology of insulin production.
concept relations (Madsen et al., 2002).
Terminological ontologies are not strictly
speaking formal ontologies but may be transformed
into such. The graph in Figure 1 is thus a
transformation into concept feature structures of the
concepts in Figure 2.
In the generative ontology, the concepts are
represented by concept feature structures, such as for
example: secretion[RES: insulin]. In terminological
ontologies the concepts are represented by terms
(linguistic expressions), e.g. insulin secretion, and
feature specifications expressing characteristics of
the concept are introduced on each concept, e.g.:
RESULT: insulin on the concept insulin secretion.
On the basis of concept relations and characteristics,
the concept feature structures of the generative
ontology may be generated, e.g. the concept insulin
secretion has the superordinate concept secretion
and the characteristic RESULT: insulin resulting in
the concept feature structure: secretion[RES:
insulin]. The two representations are closely
interlinked in that any terminological representation
can be translated into a generative representation
expressing the semantic content for each concept in
the terminological ontology.
SIABO - Semantic Information Access through Biomedical Ontologies
173

3.2 Ontology of Insulin Production
In Figure 2, we present an extract of one of the
resulting ontologies. Boxes with text in capital
letters represent subdivision criteria, the other boxes
represent concepts. The lines without arrows
represent isa relations, the arrow lines represent
other relations. Characteristics are given in the form
of feature specifications below the concept boxes.
Based on an analysis of the characteristics of the
concepts stimulation and inhibition, these concepts
were grouped under the subdivision criterion
INFLUENCE, where the distinct characteristics
clearly show the difference between them. Where
appropriate, concepts have been mapped to UMLS
in order to obtain one coherent ontology.
4 EXTRACTION OF CONCEPTS
FROM TEXT
The SIABO project investigates two approaches to
concept extraction in parallel: a synthesis-approach
relying on the generative ontology, and a pattern-
based approach which relies on knowledge extracted
from a variety of lexical resources. The two
approaches are described below.
4.1 The Synthesis Approach
The computerised text analysis employed in this
approach is conducted chiefly by the generative
ontology assisted by conventional grammars.
Ideally, a sentence is turned into one concept feature
structure in the generative ontology, which is
supposed to represent the ontological meaning
content of the sentence. This is in contrast to other
approaches to the characterization of propositional
content which take into account determiners,
negations, and logical conjunctions (e.g., Nirenburg
and Raskin, 2004).
The ontology-driven rather than syntax-driven
text processing is performed by a so-called
‘ontograbber’, which, in principle in a top-down
manner, generates feature structures to be matched
against the target sentence in the text. However,
since, in general, parts of sentences have to be
skipped as unrecognisable, the current ontograbber
prototype conducts a bottom-up analysis for
synthesising onto-terms according to the generative
ontology. In addition, the ontograbber is guided by
conventional grammar rules. However, many
potential syntactical analyses brought about by
structural ambiguities, are never actualised since
they are dismissed as category mistakes by the
ontological affinities specified on the set of
ontological relations.
In this synthesis approach, adjectives and
prepositional phrases give rise to feature structure
contributions to be attached to the concept coming
from the head noun in noun phrases. Verbs are dealt
with by nominalisation. Crucially, the ontograbber
admits partial, incomplete analysis, which in the
worst case falls back on keywords found in a
sentence and being present in the generative
ontology.
4.2 The Pattern-based Approach
In parallel with the synthesis approach, we explore a
pattern-based approach to concept extraction. Like
the synthesis approach, this approach allows us to
match phrases in text with a view to mapping the
conceptual content of these fragments into the
generative ontology. The patterns are generated from
information available from existing lexical
ressources, currently the nominalisation lexicon
NOMLEX-plus, the verb-lexicon VerbNet and
WordNet. These resources provide syntactic
argument realization rules for verbs and their
arguments and nominalised forms of verbal
expressions with semantic information in the form of
semantic roles.
The patterns form part of a process which generates
concept feature structures serving as an index for a
given text. We thereby move from string or word-
based indexing to semantics-based indexing.
As mentioned, this approach includes
nominalisation of relations expressed by verbs. We
use this nominalisation strategy for an important
reason: The set of well-formed feature structures in
the generative ontology is determined by the set of
atomic concepts in the skeleton ontology and the
chosen set of relations. In order to construct a
conceptual feature structure, we need at least one
semantic relation. However, there is no natural upper
limit to the number of possible semantic
relations. Our claim is that conceptual indexing will
lead to better results when using a small number of
semantic relations. The primary purpose of
conceptual indexing is to permit retrieval by way of
matching descriptions of queries against descriptions
of text. An increased number of semantic relations
will lead to an increased number of possible concept
descriptions. Due to the unavoidable imprecision in
the concept extraction, this increased number of
possible content descriptions will in turn lead to a
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
174

reduced probability of match between descriptions
for conceptually similar but lexically or syntactically
dissimilar linguistic expressions, and thereby lead to
a decreased recall. Thus, we need a set of relations
to express semantics, but we aim at keeping this set
at a manageable size in order to obtain the best
possible match.
5 QUERYING INFORMATION
AND KNOWLEDGE
Given a domain ontology as shown above and a set
of documents in which concepts have been
identified, the task is to provide means for query
interpretation and evaluation that draws on
conceptual content and exploits the conceptualis-
ation in the ontology.
In the present approach, query evaluation relies
on comparison of a conceptual description of the
query with conceptual descriptions of texts from the
database. A conceptual description is a set of
conceptual feature structures providing a mapping
from the text or the query to the ontology. Search in
a text collection indexed by concepts can employ
concept similarity-measures so that conceptual
reasoning can be replaced by simple similarity
computation, thereby allowing for a scaling to very
large information bases. Thus, a major challenge is
to define conceptual description similarity in terms
of the structure and relations in the ontology.
One obvious way to measure similarity in
ontologies is to evaluate the distance in the graphical
representation between the concepts being
compared, where shorter distance implies higher
similarity. A number of different ontological
similarity measures have been proposed along these
lines, for instance, Shortest Path Length (Rada,
1989), Information Content (Resnik, 1999), see also
(Budanitsky and Hirst, 2006).
An essential part of document querying is to
establish a mapping that, given a description for the
query, indicates matching – or similar – descriptions
for texts. One option is to let similarity reflect the
skeleton ontology by deriving it from the syntactic
derivation relation for conceptual feature structures,
where longer derivation paths correspond to smaller
degree of similarity. However, the comparison of
conceptual descriptions should not be merely
syntactic. Rather, description resemblance can be
measured in terms of similarity derived from all
concept relations in the ontology. Initially, in the
processing of a query, a description is generated.
Then this query description is compared, in
principle, to every conceptual description of every
document appearing in the database. Finally,
documents are ranked by the degree to which their
respective descriptions resemble the conceptual
description of the query. The query answer is a
ranking of the documents that are most similar to the
query.
In a framework where the domain of texts is
reflected in a knowledge base, as comprised by the
ontology, obviously not only the texts, but also the
domain ontology may in some cases be the target of
interest for queries. Knowledge about existence of
concepts, how concepts are related and about
similarities between concepts is also relevant. In
addition knowledge about the actual content of texts
can be viewed through the ontology simply by
means of revealing only concepts that exist in the
texts. In other words, the ontology plays a specific
role here, since it constitutes the means by which we
can obtain a conceptual view of the texts content.
Thus as an additional functionality, the user may
browse the generative ontology directly and then
follow the links to the relevant text parts by
descending to an ontological level of specialisation
with a manageable number of links to the target text.
6 CONCLUSIONS
We have presented an approach to representing,
organizing, and accessing conceptual content of
biomedical texts using a formal ontology. In
particular, we have presented the key ideas
addressing exploitation of ontologies for carrying
out content-based text search within a scientific
domain recognising not only synonyms but also
more general paraphrasations. Presently, we have
working prototypes. However, the viability of the
approach remains to be validated on a large scale, in
particular whether the devised ontological text
processing prototypes afford a
significant improvement compared with
conventional keyword search.
REFERENCES
Andreasen, T., Fischer Nilsson, J., 2004. Grammatical
Specification of Domain Ontologies. In Data &
Knowledge Engineering, 48, p. 221-230
Budanitsky, A., Hirst, G., 2006. Evaluating WordNet-
based measures of semantic distance. Computational
Linguistics, 32(1).
SIABO - Semantic Information Access through Biomedical Ontologies
175

Jensen, P., Fischer Nilsson, J., 2006. Ontology-based
Semantics for Prepositions. In Syntax and Semantics
of Prepositions, Text, Speech and Language
Technology, Vol. 29. Springer.
Ben-Avi, G., Francez, N., 2004. Categorial Grammar with
Ontology-refined Types. In Proceedings of CG04.
Fillmore, C. J., 1968. The Case for Case. In Universals in
Linguistic Theory. Holt, Rinehart, and Winston. New
York, pp. 1-88.
Hearst, M., 1992. Automatic acquisition of hyponyms
from large text corpora. In Proceedings of the 14th
International Conference on Computational
Linguistics, Association for Computational
Linguistics, Volume 2, pp. 539 – 545.
Madsen, B. N., Pedersen, B. S. & Thomsen, H. E., 2002.
Semantic Relations in Content-based Querying
Systems: a Research Presentation from the OntoQuery
Project. In Ontologies and Lexical Knowledge Bases.
Proceedings of the 1
st
International Workshop,
OntoLex 2000. OntoText Lab., Sofia, pp. 72-82.
Madsen, B.N., Thomsen, H.E., and Vikner, C., 2005.
Multidimensionality in terminological concept
modelling. In: Terminology and Content Development,
TKE 2005, 7th International Conference on
Terminology and Knowledge Engineering,
Copenhagen: 161-173.
Nilsson, J. F., 2001. A Logico-Algebraic
Framework for Ontologies. ONTOLOG.
In Ontology-based Interpretation of Noun Phrases,
Proceedings of the First Inernational. OntoQuery
Workshop, Kolding.
Nirenburg, S., Raskin, V., 2004. Ontological Semantics.
MIT Press.
Rada, R. & Bicknell, E., 1989. Ranking documents with a
thesaurus. Journal of the American Society for
Information Science, 40(5).
Resnik, P., 1999. Semantic Similarity in a Taxonomy: An
Information-Based Measure and its Application to
Problems of Ambiguity in Natural Language. Journal
of Artificial Intelligence Research.
KEOD 2009 - International Conference on Knowledge Engineering and Ontology Development
176
