
DETECTION
OF OBSTRUCTIVE SLEEP APNEA FROM THE
FREQUENCY ANALYSIS OF HEART RATE VARIABILITY
Abraham Otero
1
, Xos
´
e Vila
2
, Francisco Palacios
3
and Francisco J. Coves
3
1
Department of Software and Knowledge Engineering, University San Pablo CEU, 28668 Madrid, Spain
2
Department of Computer Science, University of Vigo, Campus As Lagoas s/n, 32004 Ourense, Spain
3
Hospital Universitary Hospital of Elche, Partida Huertos y Molinos, s/n, Elche, 03203, Spain
Keywords:
Obstructive sleep apnea (OSA), Biosignal processing, Heart rate variability.
Abstract:
This paper presents a new algorithm for the detection of Obstructive Sleep Apnea (OSA) from a single elec-
trocardiogram lead. It is based on the alterations that OSA patients present in the LF and HF bands of the
heart rate variability power spectrum. The algorithm calculates the power of the spectrum in two bands that
roughly corresponding with the LF and HF bands. Then the ratio between the power of the low band and the
power of the high band is obtained. If this ratio is greater than a certain threshold the patient is classified as
having OSA, otherwise he/she is classified as not having OSA. Then the algorithm was validated over the test
data set of the Apnea-ECG Database, classifying correctly 29 of 30 recordings.
1 INTRODUCTION
Obstructive Sleep Apnea (OSA) is a frequent sleep-
breathing disorder characterized by the presence of
total and/or partial cessations of respiratory airflow
while the patient is sleeping (K
¨
oves, 1999). These
cessations –called apneas if they are total, and hy-
popneas if they are partial– are usually caused by a
collapse in the upper airway and they produce a dis-
ruption of the patient’s sleep architecture diminishing
the refreshing effects of nocturnal rest. The overall
effect is a decrease in the patient’s quality of life, and
an increase in mortality and morbidity. OSA has a
high prevalence –it is estimated to affect 4% of male
adults and 2% of female adults – and it is recognized
as an important public health issue.
The gold standard for the diagnosis of OSA is
based on an analysis of a polysomnogram, a record-
ing of a wide range of physiological parameters ob-
tained while the patient is asleep. These sleep stud-
ies are expensive because they require the patient to
spend a night in a Hospital Sleep Unit, which must
be equipped with sophisticated and expensive elec-
tronic systems, as well as a dedicated staff. Hence
there is an interest in developing reliable diagnostic
techniques based on a smaller number of physiolog-
ical parameters that do not require dedicated Sleep
Units. The 2000 Computers in Cardiology Challenge
encouraged the development of such techniques. It
consisted of developing an algorithm capable of clas-
sifying patients as OSA or non-OSA from the modi-
fied lead V2 of the ECG (Penzel et al., 2002). For this
purpose, a database of polysomnographic recordings -
the Apnea-ECG Database- was made available to the
participants of the competition (Penzel et al., 2000).
Several clinical studies have shown that patients
with moderate-to-severe sleep apnea present alter-
ations on the heart rate variability (HRV) spectrum
(Narkiewicz et al., 1998). OSA patients present in-
creased low frequency (LF) spectral power and de-
creased high frequency (HF) spectral power. There-
fore, it is no surprise that most of the 2000 Chal-
lenge participants took advantage of information aris-
ing from the spectral analysis of the HRV (Penzel
et al., 2002). One of the simplest, but effective, al-
gorithms was the one presented by Drinnan et al.
(Drinnan et al., 2000). This algorithm calculates the
power of the HRV spectrum in two different bands,
one between 0.01 and 0.05 Hz, and another between
0.005 and 0.01 Hz. The first one corresponds ap-
proximately with the LF band, where patients suffer-
ing from OSA present increased activity. The second
band serves as a reference level: the algorithm calcu-
lates the ratio between the power of the first band and
the power of the second one, and it checks if this value
is greater than a certain threshold. If so, the patient
359
Otero A., Vila X., Palacios F. and J. Coves F. (2010).
DETECTION OF OBSTRUCTIVE SLEEP APNEA FROM THE FREQUENCY ANALYSIS OF HEART RATE VARIABILITY.
In Proceedings of the Third International Conference on Bio-inspired Systems and Signal Processing, pages 359-362
DOI: 10.5220/0002590503590362
Copyright
c
SciTePress

is classified as OSA. This algorithm correctly clas-
sified 90% of the training and test recordings of the
Apnea-ECG Database. Despite its relative success,
Drinnan’s algorithm only takes advantage of OSA pa-
tients presenting increased activity in the LF band and
the band’s limits were chosen based on a visual in-
spection of the recordings’ spectrum. Thus, it may be
possible to improve these results by tuning the bands’
boundaries and by merging information arising from
both LF and HF bands.
This paper presents a new algorithm for classify-
ing patients as OSA or non-OSA from a single ECG
lead. This algorithm is based on the alterations of the
heart rate variability that OSA patients present. Sec-
tion 2 presents the algorithm which enables patient
classification. The results of the algorithm validation
over the Apnea-ECG Database are presented in Sec-
tion 3. Finally, the results obtained are discussed and
a series of conclusions on the paper are given.
2 MATERIALS AND METHODS
After heartbeat detection, the RR intervals were fil-
tered, in order to eliminate the effects of false pos-
itives and false negatives during heartbeat detection.
Then they were resampled at 4 Hz. Cubic splines
were used for the signal interpolation; a linear inter-
polation would have produced more distortion –low
pass filter effect– than the cubic alternative (Vila et al.,
1997). The filtered and resampled RR intervals were
stored in ASCII files.
OSA patients have an increased activity in the LF
band of the HRV power spectrum, and a decreased
activity in the HF band. Thus the ratio between the
power of the LF band and the power of the HF band
power should be larger for OSA patients than for con-
trol patients. This is the idea behind our algorithm.
Therefore, we need to find the limits of both bands,
and a threshold that allows us to distinguish between
OSA and control patients.
A Matlab script that takes as inputs the lower and
upper limits of two bands corresponding with the LF
and HF regions was created. The script loads the
RR distances from the ASCII files and it calculates
the spectrum of the complete RR intervals time series
corresponding with each of the 10 normal patients.
For each patient it calculates the ratio between the
power of the LF band and the power of the HF band.
The maximum of these values is considered as the
boundary between control and OSA patients. Thus,
it is used as the threshold for the classification of the
20 OSA patients of the training set. If the ratio be-
tween the two spectrum bands of a class A recording
is greater than the threshold, the patient is classified
as OSA. Otherwise, he/she is normal. After its exe-
cution, the script reports the numbers of correctly and
incorrectly classified recordings.
Given certain values for the limits of the LF and
HF bands, they will be more suitable for our purpose
the more class A recordings they classify correctly. A
second Matlab script was created to provide the bands
to the first script, and to analyze its output. This script
starts with two initial bands selected on the basis of
a visual inspection of the spectrum of the 30 train-
ing set recordings. The initial limits for the low band
were set between 0.01 and 0.05 Hz, and for the high
band between 0.18 and 0.28 Hz. This script gradually
changes the lower and upper limits of the bands by
increasing and decreasing their values.
This second script performs an exhaustive search
looking for the optimal classification bands. For each
band limit modification, it invokes the first script and
stores the number of recordings correctly classified,
as well as the bands used in this classification. Once it
has checked all the preprogrammed bandwidths, it an-
alyzes the results and presents a list of the best bands
that have been found. This script ran for two days on
a PC with an Intel Core 2 Duo processor at 2.2 GHz
and 4 GB of RAM. The results obtained suggested
that the best classifications were obtained when the
upper limit of the low band was close to or equal to
the lower limit of the high band, and they both had a
value of approximately 0.08 Hz.
The scripts were modified in order to force the up-
per limit of the low band to always be equal to the
lower limit of the high band. The initial values of
the bands were also modified to be close to the val-
ues with which the best results were obtained in the
first run. Finally, we decreased the magnitude of the
increases and decreases that would be applied to the
lower and upper limits of each band. This time the
initial limits for the low band were set between 0.02
and 0.08 Hz, and for the high band they were set be-
tween 0.08 and 0.30 Hz, and the script applied finer
modifications to the bands. After running the script
for one day, we found that by setting the lower limit
of the low bandwidth between 0.021 and 0.048 Hz;
the upper limit of the low band (and the lower limit of
the high band) between 0.056 and 0.078 Hz; and the
upper limit of the high band between 0.232 and 0.276
Hz, all the control recordings and 28 of the 30 OSA
recordings were correctly classified.
To select the final frequencies, we looked for the
bands which maximize the separability of the class A
and the class C test set recordings. To this end, we
subtracted the threshold used in the classification; i.e,
the maximum of the ratios obtained for the class C
BIOSIGNALS 2010 - International Conference on Bio-inspired Systems and Signal Processing
360
recordings, from the LF/HF ratios obtained for each
of the class A recordings, and we calculated the sum
of all these values. This value can be considered as a
measure of the separability of the class of the control
patients and the class of the OSA patients: the higher
this value is, the greater the average distance from the
OSA patients to the control patient which is “closest”
to the OSA class is, according to this metric.
The final bands were selected in such a way that
they maximize this distance. The low band was set be-
tween 0.026 and 0.06 Hz, and the high band between
0.06 and 0.25 Hz. These bands yield a threshold value
of 0.42349; i.e., the maximum of the ratios between
the power of the low band and the power of the high
band for the 10 control patients was 0.42349. This
value will be the boundary between classes A and C.
The optimal classification bands we have found
differ significantly from the values traditionally asso-
ciated with the LF and HF bands. We tried to classify
the recordings using bands as close as possible to the
traditional LF and HF bands (between 0.04 and 0.14
Hz, and between 0.14 and 0.4 Hz, respectively). To
this end, we launched our training script with these
initial values for the bands’ boundaries and we al-
lowed only small changes to be made. In the best
case obtained, 6 training set and 8 testing set record-
ings were misclassified. These results suggest that the
bands in which patients suffering from OSA present
HRV alterations do not exactly match the traditional
definitions of the LF and HF bands.
3 RESULTS
Using the bands selected in the training phase, we
validated our algorithm using the test set recordings
of the Apnea-ECG Database. Before applying the
algorithm, the threshold value was rounded to 0.43.
This change did not affect the classification results
obtained for the training set recordings. All patients
in the control test set were correctly classified as non
OSA. Of these recordings, the one which provided
the bigger LF/HF ratio value, i.e, the one which was
“closest” to the OSA class, was 17c, which yielded a
value of 0.41. Among the patients who suffered OSA,
only one was classified as healthy (the 15a); all others
were correctly classified.
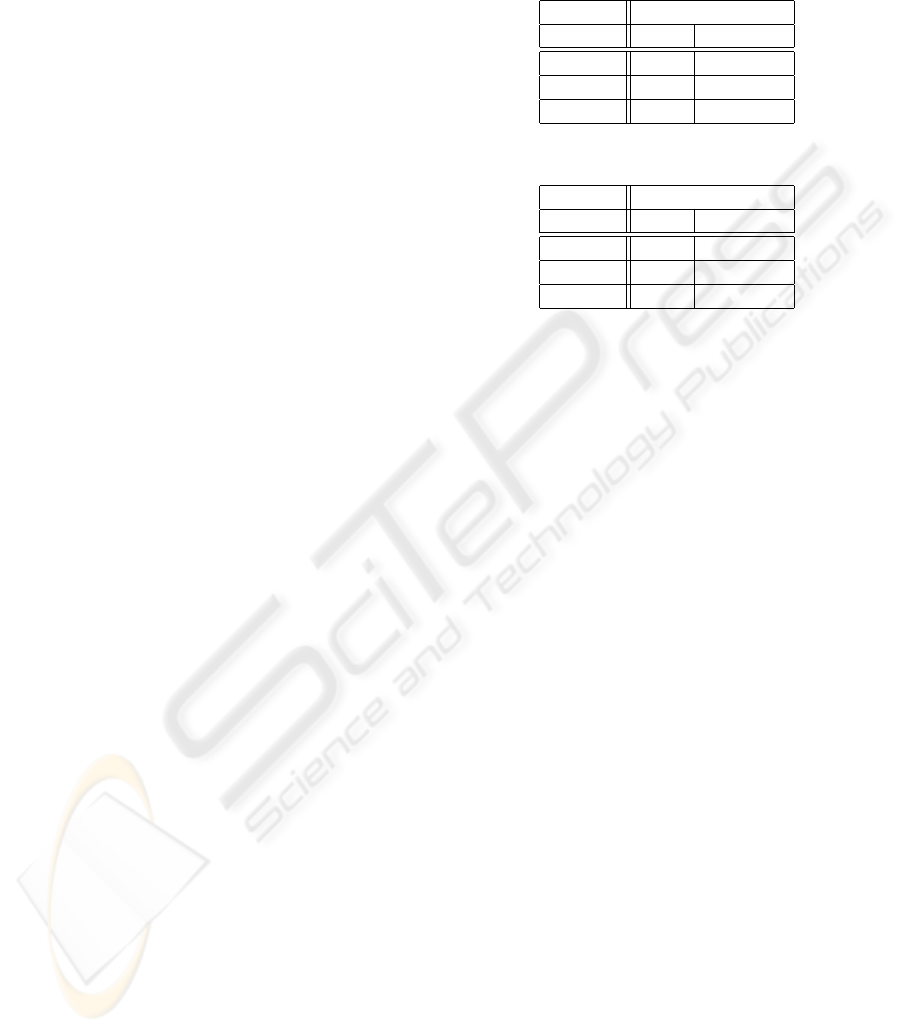
Tables 1 and 2 summarize the results obtained
over the training and test sets, respectively. These ta-
bles also show the classification results for the five
Class B recordings of the training and test sets. Given
that these recordings correspond to patients who are
on the borderline between normality and suffering
from the disorder, it is not possible to use them to
judge the quality of the results of a binary classifier as
the one proposed here.
Table 1: Training set recordings classification results.
Class Classification
OSA non-OSA
Class A 18 2
Class B 3 2
Class C 0 10
Table 2: Test set recordings classification results.
Class Classification
OSA non-OSA
Class A 19 1
Class B 2 3
Class C 0 10
4 DISCUSSION
Our algorithm correctly classifies 95% of the 60 OSA
and control recordings of the Apnea-ECG Database,
compared to 90% of the Drinnan et al. algorithm that
inspired ours. This has been achieved by taking ad-
vantage of the altered HRV that OSA patients present
both in the LF band and in the HF band and by in-
cluding a semi–automatic training stage to fine tune
the bands’ boundaries. All the classification errors
committed by our algorithm, and by the Drinnan et al.
algorithm, correspond to OSA patients who are mis-
classified as normal. From the viewpoint of the con-
struction of a diagnostic or a screening test, the classi-
fication of a patient who suffers from OSA as healthy
is less desirable than the classification of a healthy
patient as having OSA. Therefore, we might prefer an
algorithm that, even though it incorrectly classifies a
higher number of patients, it classifies less OSA pa-
tients as healthy.
To achieve this in the training set, we would have
had to set the threshold used by our algorithm to 0.32.
In the case of the test set, the threshold would have
had to have been set to 0.35014. In the first case, this
would lead to the classification of 6 of the 10 con-
trol patients as having OSA; in the latter 5 control pa-
tients would have been misclassified. An algorithm
that classifies half or more of the healthy patients as
having OSA does not seem a good compromise.
A visual inspection of the 3 incorrectly classified
recordings shows that they do not present altered HRV
in the LF or HF bands. Visual inspection also reveals
that some recordings only present alterations in one
of the two bands. In these cases, our algorithm is still
DETECTION OF OBSTRUCTIVE SLEEP APNEA FROM THE FREQUENCY ANALYSIS OF HEART RATE
VARIABILITY
361

able to make the correct classification thanks to the
merging of information arising from both bands. In
the case of the Drinnan et al. algorithm, the patient
must present altered activity in the only band the al-
gorithm considers –LF.
The fact that neither the Drinnan et al. algorithm
nor ours classified any of the control patients as hav-
ing OSA suggests that both algorithms have a rather
high specificity. This specificity may have its root in
the fact that healthy patients do not usually show al-
tered HRV activity in any of the two bands, although
some OSA patients do not present the typical altered
activity, despite their disorder.
The scientific literature states that patients suffer-
ing from OSA have decreased activity in the LF band
and increased activity in the HF band. However, the
best classification bands found by our scripts for the
Apnea-ECG Database recordings differ from the tra-
ditional LF and HF bands. Our high band –between
0.06 and 0.25 Hz– incorporates a fragment of the LF
band (between 0.06 and 0.014), and our low band –
between 0.026 and 0.06 Hz– includes only the lower
part of the LF band and the VLF band (Very Low Fre-
quency, between 0.002 and 0.004 Hz).
However, the algorithm is based on patients suf-
fering from OSA having less power in the high band
and more power in the low band, because it calculates
the ratio between the low band and the high band, and
expects that OSA patients have a higher value for this
ratio than healthy patients. Furthermore, in our at-
tempt to find bands as close as possible to the LF and
HF bands, the classification results are significantly
worse than those obtained with the optimal bands –
6 recordings misclassified in the training set, and 8
in the testing test. Therefore, our results suggest that
OSA patients have increased activity only in the lower
part of the LF band and in the VLF band, and have de-
creased activity not only in part of the HF band, but
also in the 0.06 and 0.014 Hz range of the LF band.
5 CONCLUSIONS
This paper presents a simple but effective algorithm
capable of classifying patients as OSA or non-OSA
on the basis of a single ECG lead. The algorithm cal-
culates the power spectrum of the time series obtained
by filtering and resampling at 4 Hz the RR intervals
extracted from the ECG lead. Then the power of the
bands between 0.026 and 0.06 Hz and between 0.06
and 0.25 Hz is calculated, and then the ratio between
the power of the low band and the high band is ob-
tained. If this ratio is greater than a certain threshold
–0.43– the patient is classified as OSA. The bound-
aries of the bands and the threshold were obtained
by means of a semi-automatic training stage where
the Apnea-ECG Database training data set was used.
The algorithm was validated on the test data set of
the same database, incorrectly classifying only 1 of
30 control and OSA recordings.
Our future work aims to obtain a screening test
for OSA patients that can be performed at low cost
and in the patient’s home. This can avoid travels to
the hospital, long waiting lists, and other inconve-
niences for the patient. To this end, the test will be
based on MEDIM, a PDA platform with the capabil-
ity of recording ECGs (Presedo et al., 2009). The
algorithm we have developed has a low demand of
computational resources, which makes it suitable to
be implemented in such a device.
ACKNOWLEDGEMENTS
This work was supported by the Spanish MEC and the
European FEDER under the grant TIN2006-15460-
C04-02.
REFERENCES
Drinnan, M., Allen, J., Langley, P., and Murray, A. (2000).
Detection of sleep apnoea from frequency analysis
of heart ratevariability. In Computers in Cardiology
2000, pages 259–262.
K
¨
oves, P. (1999). Obstructive Sleep Apnea Syndrome.
Springer Verlag.
Narkiewicz, K., Montano, N., Cogliati, C., van de Borne,
P., Dyken, M., and Somers, V. (1998). Altered cardio-
vascular variability in obstructive sleep apnea. Circu-
lation, 98:1071–1077.
Penzel, T., McNames, J., de Chazal, R., Raymond, B., Mur-
ray, A., and Moody, G. (2002). Systematic compari-
son of different algorithms for apnea detection based
on electrocardiogram recordings. Med. Biol. Eng.
Comput, 40:402–407.
Penzel, T., Moody, G., Mark, R., Goldberger, A., and Peter,
J. (2000). The apnea-ecg database. In Computers in
Cardiology 2000, volume 27, pages 255–258.
Presedo, J., Tarasc
´
o, M., Teijeiro, T., F
´
elix, P., Castro, D.
(2009). A PDA-based modular and multipurpose sys-
tem for intelligent ubiquitous monitoring. In WISP
2009, pages 223–228.
Vila, J., Palacios, F., Rodr
´
ıguez-Presedo, J., Fernandez-
Delgado, M., F
´
elix, P., and Barro, S. (1997). Time-
frequency analisys of herat-rate variability. IEEE Eng.
in medicine and biology magazine, 16:119125.
BIOSIGNALS 2010 - International Conference on Bio-inspired Systems and Signal Processing
362
