
SYSTEMATIC POSITION AND PHYLOGENETIC
RELATIONSHIPS OF THE CYCLOPHYLLIDEAN CESTODES
An In-silico Study using ITS2 rDNA and Sequence-structure Alignment
Veena Tandon, Devendra Kumar Biswal
Department of Zoology, North-Eastern Hill University,Shillong, Meghalaya, India and Bioinformatics Centre
North-Eastern Hill University, Meghalaya, India
Pramod Kumar Prasad
Department of Zoology, North-Eastern Hill University,Shillong, Meghalaya, India
Chenkual Malsawmtluangi
Department of Zoology, North-Eastern Hill University,Shillong, Meghalaya, India
Keywords: Mesocestoides, Cyclophyllidea Cestodes, ITS2 rDNA, Molecular Morphometrics, Sequence-Structure
alignment.
Abstract: The phylogenetic relationships and systematic position of cyclophyllidean cestodes have always been
controversial and opinions of different authors on the systematic rank and content of this order have varied
greatly. Molecular phylogenetic analysis based on ITS2 rDNA of 16 representatives spanning 6 different
families (Mesocestoididae, Davaineidae, Anoplocephalidae, Taeniidae, Dipylidiidae and Hymenolepididae)
of the Order Cyclophyllidea and one out group from the family Diphyllobothriidae of the Order
Pseudophyllidea confirmed the monophyletic nature of the Order Cyclophyllidea. Further, the results were
validated by bayesian analysis, primary sequence-structure alignment and subsequent molecular
morphometrics analysis. At the major nodes all the trees from various analyses were similar. Mesocestoides
was interestingly accommodated within Cyclophyllidea and served as a sister clade close to the families
Taeniidae, Anoplocephalidae, Hymenolepididae and Dipylidiidae.
1 INTRODUCTION
Cyclophyllidean cestodes are parasites occurring as
adult or larval stage in a wide variety of avian and
mammalian hosts and are cosmopolitan in nature
commonly called tapeworms that live in the
digestive tract of vertebrates as adults and often in
the bodies of various animals as juveniles.
Taxonomists face inconsistent or even contradictory
information when they examine the systematic
relationships between cestodes at higher taxonomic
groupings (Mariaux, 1999). The phylogeny at higher
levels is of little significance as the taxonomy is
primarily based on morphological characters and in
case of cestodes it is often difficult to discern
between secondary loss and convergence of
morphological characters as several authors have
weighed the characters of taxa differently (Khalil et
al., 1994). This applies well to the taxonomic
positions of Mesocestoides, which is quite
complicated by a high degree of non-significant
morphological variation. Cyclophyllidean cestodes
of the Family Mesocestoididae differ from other taxa
in the Order Cyclophyllidea in some important
characteristics. The life cycle of Mesocestoides spp.
requires three hosts and not two; the median ventral
position of the genital atrium and the presence of
bipartite vitelline gland in Mesocestoides spp. appear
to be unique among the Cyclophyllidea (Chandler,
1946).
Although, nowadays, most authors agree that
there are 15 families included in the monophyletic
Order Cyclophyllidea, till date no study has
pinpointed the complete taxonomic linkage of all the
5
Tandon V., Kumar Biswal D., Kumar Prasad P. and Malsawmtluangi C. (2010).
SYSTEMATIC POSITION AND PHYLOGENETIC RELATIONSHIPS OF THE CYCLOPHYLLIDEAN CESTODES - An In-silico Study using ITS2 rDNA
and Sequence-structure Alignment.
In Proceedings of the First International Conference on Bioinformatics, pages 5-12
DOI: 10.5220/0002690500050012
Copyright
c
SciTePress

15 families persuasively with genetic evidence.
Besides, the taxonomic position of Mesocestoididae
remains unclear as some of the cestodes at the
generic level of the family bear similarity to those of
the Order Pseudophyllidea; one such is the case of
M. lineatus, with a wide range of hosts thus lacking
host specificity, which is atypical of Cyclophyllidea
but resembles the cestodes (viz. Diphyllobothrium
dendriticum) of the Order Pseudophyllidea
(Kamegai et al., 1967).
The phylogeny of cyclophyllidean cestodes has
been reported with aid of 12S rRNA markers of
mitochondrial genomic regions from 21 cestode
species spanning eight families (von Nickisch-
Rosenegk et al., 1999). In the present study, we
address the overall taxonomic resolution of
cyclophyllidean cestodes with a different
phylogenetic marker using a combinatorial approach
of sequence analysis and molecular morphometrics.
The internal transcribed spacer 2 (ITS2), the region
of ribosomal RNA between 5.8S rRNA gene and the
large subunit (28S rRNA) has proven to be
appropriate marker for analysis of microscale
phylogenies of close relatives (Coleman, 2003).
Moreover, the ITS2 sequence data can be subjected
to secondary structure predictions and as the
secondary structure seems to be well conserved, it
can provide clues for higher taxonomic studies
(Schultz et al., 2005). This is quite obvious that
phylogenetic analyses are improvable by inclusion
of molecular morphometrics information in
common sequence analysis (Billoud et al., 2000).
Here, we combine sequence with structural
information and apart from the biological problem,
address the different in-silico practices in vogue for
phylogeny studies using ITS2 r-DNA.
2 MATERIALS AND METHODS
2.1 Sequence Alignment and Molecular
Phylogenetic Analysis
The Cyclophyllidean Cestoda sequences from
several geographical locations spanning six different
families and one from the Pseudophyllidea order
(taken as out group) were retrieved from the NCBI
GenBank databases in the present study. Nucleotide
sequences were aligned and edited using ClustalW
(Thompson et al., 1994). A phylogenetic tree was
constructed using the Neighbor-Joining as well as
Maximum Parsimony methods in MEGA 4.0
(Tamura et al., 2007)). Branch support was given
using 1000 bootstrap replicates. Maximum
parsimony was accomplished with gaps treated as
missing data and all characters coded as “unordered”
and equally weighted.
2.2 Bayesian Phylogenetic Analysis
A Bayesian analysis using MrBayes V 3.12
(Ronquist and Huelsenbeck, 2003) was carried out
for tree construction using a general time reversible
substitution model (GTR) with substitution rates
estimated by MrBayes. Metropolis-Coupled Markov
Chain Monte Carlo (MCMCMC) sampling was
performed with two incrementally heated chains that
were combinatorially run for 20,000 generations.
The convergence of MCMCMC was then monitored
by examining the value of the marginal likelihood
through generations. Coalescence of substitution rate
and rate model parameters were also examined.
Average standard deviation of split frequencies was
checked and the generations were kept on adding
until the standard deviation value was below 0.01.
The values slightly differed because of stochastic
effects. The sample of substitution model parameters
and samples of trees and branch lengths were
summarized by the “sump burnin” and “sumt
burnin” commands, respectively. The values in the
following commands were adjusted as per the 25%
of our samples. A cladogram with the posterior
probabilities for each split and a phylogram with
mean branch lengths were generated and
subsequently read by the tree drawing program Tree
view V1.6.6 (Page, 1996).
2.3 ITS2 Secondary Structure
Prediction and Sequence Structure
Alignment
ITS2 secondary structures of the cestodes were
folded with the help of MFold (Zuker, 2003) by
screening for thermodynamically optimal and
suboptimal secondary structures (default settings,
with T=25
o
C). The secondary structures in Vienna
(dot-bracket-dot) format was used as an input for
MARNA (Siebert and Backofen, 2005) to calculate
sequence-structure multiple alignment. However,
there was a limitation with the online server that the
maximum length of one RNA sequence is restricted
to 500 bases; hence some of the ITS sequences
whose exact boundary information was available
from GenBank graphics view was trimmed for
facilitating MARNA to run. Some more cestode
sequences were also taken whose 5.8S, 28S and
ITS2 regions were clearly defined so as to include in
the sequence-structure multiple alignment dataset. A
phylogenetic tree was created using ProfDistS
(Wolf. et al., 2008) that takes the multiple aligned
BIOINFORMATICS 2010 - International Conference on Bioinformatics
6
sequence-structure as an input and a consensus tree
was built using RNA/DNA structure profile
neighbor-joining method with 100 bootstraps.
Besides, the GC content of the ITS 2 regions was
calculated using Oligo Calculator available at
http://www.pitt.edu/~rsup/OligoCalc.html.
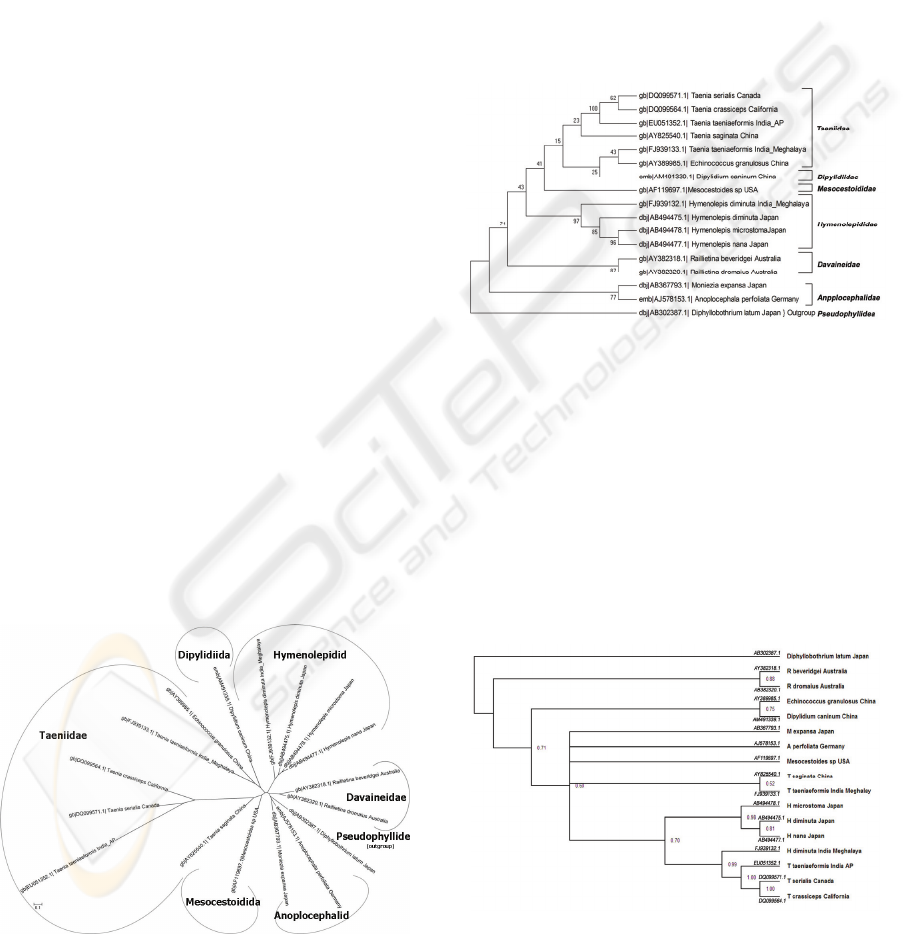
3 RESULTS
3.1 Neighbour-Joining (NJ) and
Maximum Parsimony (MP) Trees
GenBank accession numbers of ITS2 sequences for
the cestodes spanning 6 families of the Order
Cyclophyllidea and one from the Order
Pseudophyllidea (as out group) are given in Table 1.
The evolutionary history was inferred using the NJ
method (Saitou and Nei, 1987) and the bootstrap
consensus tree (Fig. 1), inferred from 1000
replicates, depicted an overall robust topology of the
cyclophyllidean cestodes’ phylogeny. Branches
corresponding to partitions reproduced in less than
50% bootstrap replicates are collapsed. The
evolutionary distances were computed using the
Maximum Composite Likelihood method (Tamura
et al., 2004) and are in the units of the number of
base substitutions per site. All positions containing
gaps and missing data were eliminated from the
dataset (complete deletion option). There was a total
of 168 positions in the final dataset.
With MP method (Dayhoff et al., 1965) the most
parsimonious tree drawn had the length=727. The
percentage of replicate trees in which the associated
taxa clustered together in the bootstrap test (1000
replicates) are shown next to the branches (Fig. 2)
Figure 1: Neighbor-Joining Tree.
and the MP tree was obtained using the Close-
Neighbor-Interchange algorithm (Nei and Kumar,
2000) with search level 3 in which initial trees were
obtained with the random addition of sequences.
After deletion of the positions containing the gaps,
there were a total of 168 positions in the final dataset
out of which 139 were parsimony informative.
The phylogenetic analysis using the distance and
character state methods showed very good bootstrap
values (Figs. 1 & 2) and all the six cyclophyllidean
families depicted reliable monophyletic groupings.
Bootstrap values for the six monophyletic groups
ranged from 70-100%.
Figure 2: Maximum Parsimony Tree.
3.2 Bayesian Analysis
Bayesian analysis of the alignment retained the same
topology and supported the branches with good
bootstrap values (Fig. 3), though there were slight
variations in the placing of some species of the
Family Taeniidae (Taenia saginata from China and
Taenia taeniaeformis from India) that were grouped
in another node from the rest of the Taenia species.
Diphyllobothrium latum of the Order
Pseudophyllidea was rooted as an out group.
Figure 3: Bayesian Analysis Phylogeny.
SYSTEMATIC POSITION AND PHYLOGENETIC RELATIONSHIPS OF THE CYCLOPHYLLIDEAN CESTODES -
An In-silico Study using ITS2 rDNA and Sequence-structure Alignment
7

Table 1: Cyclophyllidean Cestode species used in this study with the respective GenBank accession numbers for the
corresponding ITS 2 sequences. (*) Designated as outgroup.
Species GenBank Accession No. Order & Family
H
ymenolepis diminuta India_Meghalaya FJ939132.1 Cyclophyllidea: Hymenolepididae
H
ymenolepis diminuta Japan AB494475.1 Cyclophyllidea: Hymenolepididae
H
ymenolepis microstoma Japan AB494478.1 Cyclophyllidea: Hymenolepididae
H
ymenolepis nana Japan AB494477.1 Cyclophyllidea: Hymenolepididae
D
ipylidium caninum China AM491339.1 Cyclophyllidea: Dipylidiidae
Taenia saginata China AY825540.1 Cyclophyllidea: Taeniidae
E
chinococcus granulosus China AY389985.1 Cyclophyllidea: Taeniidae
Taenia serialis Canada DQ099571.1 Cyclophyllidea: Taeniidae
Taenia crassiceps California DQ099564.1 Cyclophyllidea: Taeniidae
Taenia taeniaeformis India_Andhra Pradesh EU051352.1 Cyclophyllidea: Taeniidae
Taenia taeniaeformis India_Meghalaya FJ939133.1 Cyclophyllidea: Taeniidae
M
oniezia expansa Japan AB367793.1 Cyclophyllidea: Anoplocephalidae
A
noplocephala perfoliata Germany AJ578153.1 Cyclophyllidea: Anoplocephalidae
M
esocestoides spp. USA AF119697.1 Cyclophyllidea: Mesocestoididae
R
aillietina beveridgei Australia AY382318.1 Cyclophyllidea: Davaineidae
R
aillietina dromaius Australia AY382320.1 Cyclophyllidea: Davaineidae
*Diphyllobothrium latum Japan AB302387.1 Pseudophyllidea:Diphyllobothriidae
3.3 Secondary Structure Analysis and
GC Content
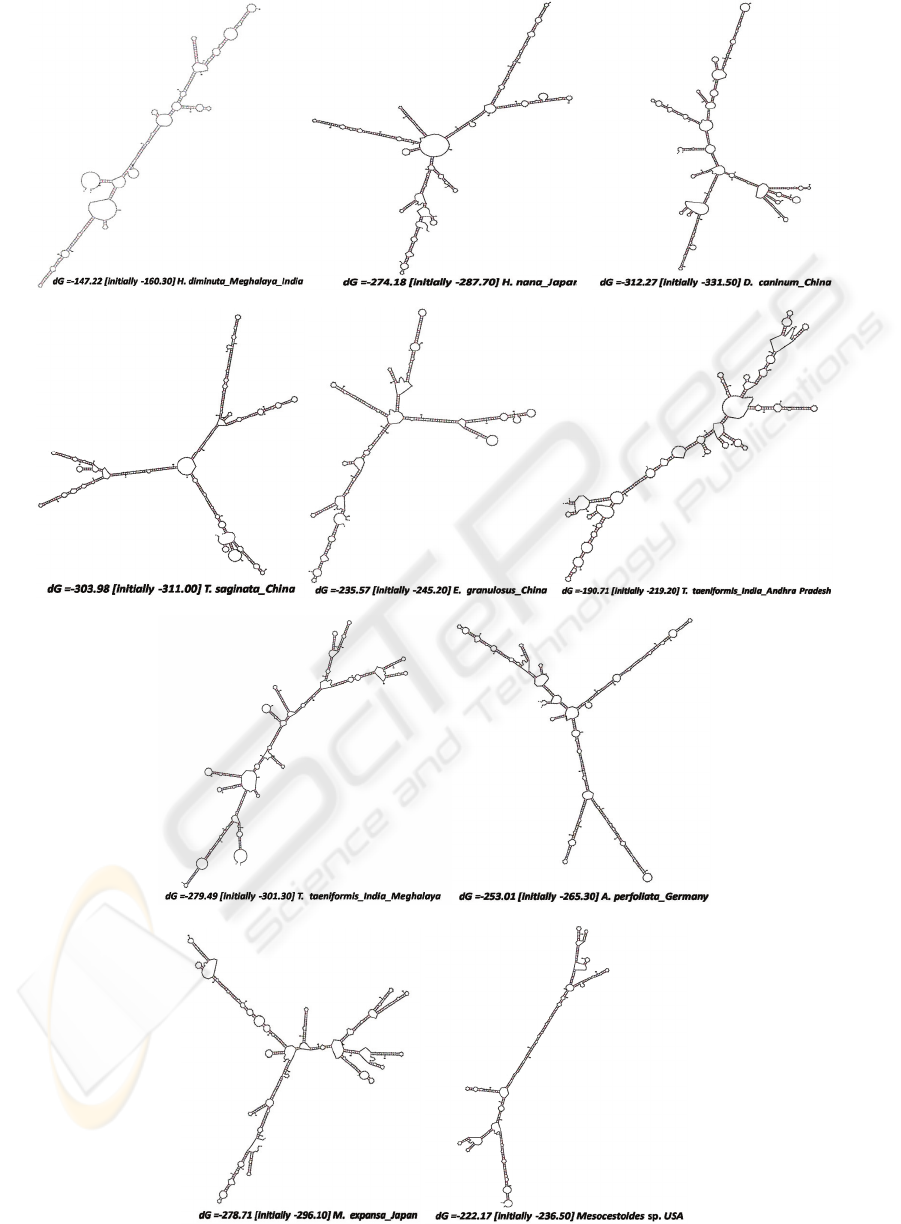
3.3.1 ITS2 Secondary Structures
The ITS 2 secondary structures (Figs. 4, 5A-I) were
analyzed for conserved stem and loop. The
Hymenolepis species showed characteristic hallmark
of ITS 2 secondary structure, i.e., four helices were
clearly visible in secondary structures with third one
as the longest. However, the third helix contains a
side branch (Fig. 4). Species of Mesocestoides and
Raillietina also maintained common secondary ITS2
core structure. Taenia and Echinococcus species
showed a lot of variation in the secondary structure
with many extra helices, loops and side branches.
UGGU motif (Fig. 4) in the secondary structure was
present in almost all the species of Hymenolepis
genus and the U-U mismatch motif was completely
absent in them. Secondary structures of species,
belonging to the same genus showed high overall
structural similarity except Taenia species in which
considerable differences were noticed. The grouping
of the families Taeniidae, Mesocestoididae and
Hymenolepididae together in phylogenetic trees
(Figs. 1, 2 & 3) forming a monophyletic group was
supported by ITS2 secondary structure similarity.
3.3.2 GC Content
The GC content in the ITS2 region was calculated
(Table. 2) and it was found that for Taenia species
the GC content varied from 54.6% to 62.6%. For
species of Hymenolepis, it ranged from 43.7% to
54%. Taenia species showed a higher GC content
compared to others. The GC content also somewhat
reflected grouping pattern of the organisms in the
phylogenetic tree. Among many other factors, GC
content is one of the factors related with stability of
the secondary structure.
Table 2: Percentage of GC content in the ITS2 region of
various cyclophyllidean cestodes.
Organism name GC Content
Taenia saginata 62.4%
Taenia serialis 62.6%
Taenia crassiceps 57%
Taenia taeniaeformis 61.6%
Taenia pisiformis 54.6%
Hymenolepis nana 50.5%
Hymenolepis diminuta 43.7%
Raillietina beveridgei 50.5%
Raillietina australis 49.4%
Mesocestoides spp. 58.8%
Anoplocephala perfoliata 46%
Echinococcus granulosus 59%
Figure 4: ITS2 Secondary Structure showing UGGU
motif: Hymenolepis nana.
UGGU
BIOINFORMATICS 2010 - International Conference on Bioinformatics
8
5A 5B 5C
5D 5E 5F
5G 5H
5I 5J
Figure 5A-J: ITS2 Secondary structures of Cyclophyllideab cestodes.
SYSTEMATIC POSITION AND PHYLOGENETIC RELATIONSHIPS OF THE CYCLOPHYLLIDEAN CESTODES -
An In-silico Study using ITS2 rDNA and Sequence-structure Alignment
9
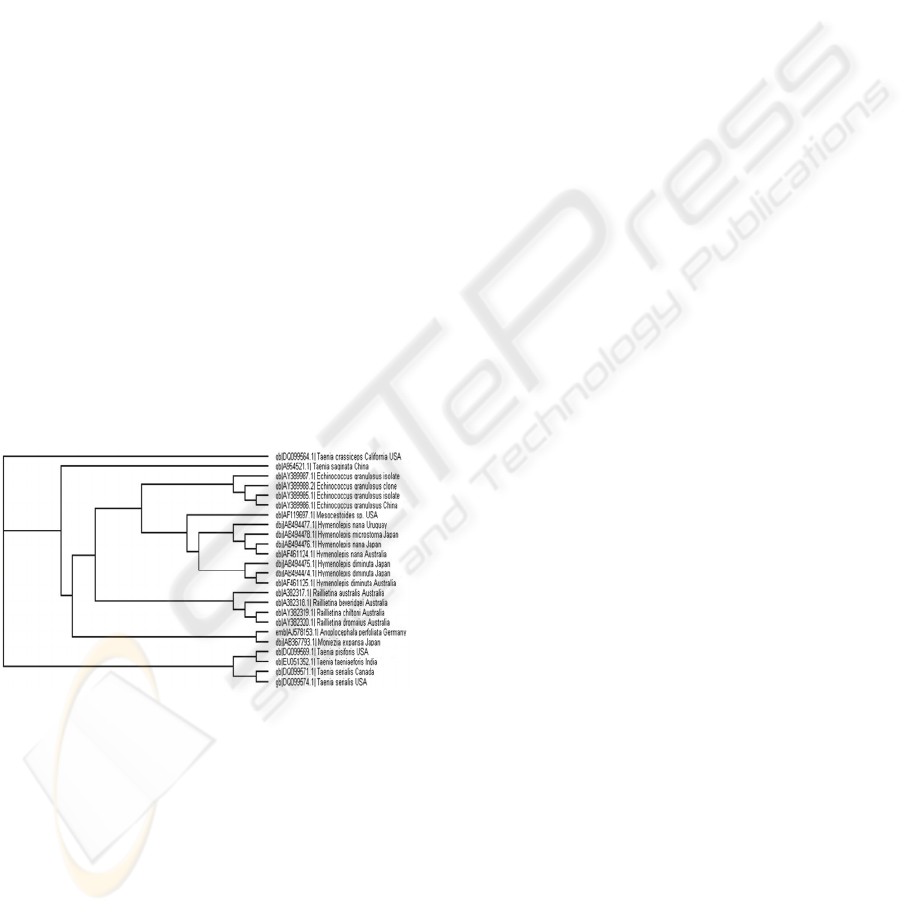
3.3.3 Primary Sequence-secondary
Structure Alignment
Apart from the ITS 2 sequences initially used for the
primary sequence analysis, some more sequences
were included for secondary structure alignment
with the primary sequence. Multiple sequence-
structure alignment from MARNA was used in
ProfDistS program to build phylogenetic tree (Fig.
6); though secondary structure information helped to
improve alignment, the proposed phylogeny showed
slight differences. However, the monophyletic
character of different groups was retained with few
exceptions. The Echinococcus species were placed
close to species of Hymenolepis. Taenia saginata
and Taenia crassiceps were placed in the upper
branch of the phylogenetic tree away from the basal
group of Taenia species. These slight differences in
tree topology may be due to specific ITS2 rate
matrix used in the analysis. As most of the studies
related to ITS2 have been carried out pertaining to
plants and fungi, the specific rate matrix developed
largely depends on those data. The ITS2 region of
cestodes may follow different rates of evolution and
thus ITS2 rate matrix specific to cestode may
provide better results. Overall there was
considerable similarity between the ITS analysis and
the consensus of previous phylogenetic
reconstruction using other DNA loci.
Figure 6: Multiple sequence-structure alignment.
4 DISCUSSION
The ITS2 region is a requisite in ribosome
biogenesis (Cote et al., 2001) and its gradual
removal from mature rRNA is driven by its specific
secondary structure. Using the secondary structure
of ITS2 sequences from various cyclophyllidean
cestodes covering six important families in this
study, we aimed to pursue three consecutive queries
concerning their systematic relationships (i) whether
the cyclophyllidean cestodes retain the same
taxonomic positions at higher levels and can be
regarded as monophyletic considering ITS2 as
molecular markers for drawing the phylogeny; (ii)
how does the secondary structure of ITS2 sequences
contribute to the Cyclophyllidea phylogeny using
molecular morphometrics approach, and whether the
latter would corroborate the monophyletic
characteristics at the family level?
The order Cyclophyllidea has 15 families. Of
these 6 cestode species belonging to families
Hymenolepididae, Dipylidiidae, Taeniidae,
Anoplocephalidae Mesocestoididae and Davaineidae
were considered for the analysis. Using the ITS2
sequence data of Diphyllobothrium latum that
represents the Order Pseudophyllidea as an out
group, we constructed phylogenetic trees using
distance-based, character-based and Bayesian
methods. Besides, molecular morphometrics
approach was employed taking sequence-structure
alignment into consideration. Our study shows that
all the taxa were clearly monophyletic within their
families and principally correspond to earlier
classifications based upon morphology and biology
(Khalil et al., 1994).
The genus Mesocestoides has a complicated
taxonomy owing to its high degree of nonsignificant
morphological variations; the genital pore is median,
scolex armature and rostellum are lacking, the ovary
and vitellaria both consist of two compact masses,
and a paruterine organ develops at the posterior end
of the tube like uterus. The Order Mesocestoididea is
placed between the Trypanorhyncha and the
Tetrabothridea (Wardle et al., 1974), while the two
known genera (Mesocestoides and Mesogyna) are
raised to family rank, with an uncertain relationship
with Cyclophyllidea (Khalil et al., 1994). The
families Mesocestoididae and Taeniidae share a
common origin as tentatively suggested on the basis
of tegumental hairs of their metacestodes, in contrast
to cysticercoids of other cyclophyllideans, have
series of fibrous layers instead (Brooks et al., 1991).
Moreover, the entire lifecycle of Mesocestoides is
quite aberrant and the number of intermediate hosts
remains enigmatic. Our dendrograms, predicted
through several in-silico approaches, demonstrate
that Mesocestoides spp from USA are closer to the
families Anoplocephalidae and Taeniidae. Due to
the lack of ITS 2 sequences of other Mesocestoidae
genera in the public domain, we could not build a
better dataset for accurate resolution of the family
with high precision. Nevertheless our data supported
Khalil et al’s (1994) arrangement of Mesocestoides
into Cyclophyllidea and a narrow relationship
BIOINFORMATICS 2010 - International Conference on Bioinformatics
10

between Mesocestoididae, Taeniidae,
Hymenolepididae and Anoplocephalidae.
Taeniids are the best-known cestodes. The
various phylogenetic methods applied to Taenia and
Echinococcus corroborates the monophyletic
grouping of the family Taeniidae. The present
analysis agrees to the monophyly of other families
under Cyclophyllidea; further analysis can be done
once more and more molecular markers are
deposited in public gene bank databases.
5 CONCLUSIONS
Molecular morphometrics approach that uses
combined features both from anatomical and
quantitative morphometrics and molecular primary
sequence comparison was the basis of our study. The
approach differentiates significant features between
anatomical and molecular characters that make
molecular morphometrics a strong predictive tool for
phylogenetic resolution. There is always more than
one gene involved in anatomical variations and most
importantly the genetic sites responsible for
morphological characters are usually not known. On
the contrary, molecular structural variations are
because of identifiable mutations that can be
characterized at the single mutational level. The
observed anatomical characters are the outcome of
both the genetic characters as well as epigenetic
effects (environmental influences) whereas the
molecular morphometrics method takes advantage of
the fact that molecular characters are independent of
their somatic expression (Smith, 1992).
The analysis corroborated strong results for
phylogenetic relationships of cyclophyllidean
cestodes and this was so because of using ITS2 data
as phylogenetic molecular markers and the inclusion
of secondary structure information that offers a
resolution power for relationships from the level of
sub species up to the order level.
ACKNOWLEDGEMENTS
This study was carried out under the DIT sponsored
project, “Northeast Parasite Information and
Analysis Centre” sanctioned to VT by the Ministry
of Communication & Information Technology
(Government of India) and Bioinformatics Centre
NEHU with partial support from UGC-supported
UPE-Bioscioences Programme in the School of Life
Sciences at NEHU.
REFERENCES
Billoud, B., Guerrucci, M. A., Masselot, M., Deutsch J.S.,
2000. Cirripede phylogeny using a novel approach:
molecular morphometrics. Molecular Biology and
Evolution, 17(10) pp. 1435-1445.
Brooks, D. R., Hoberg, E. P., Weekes, P. J., 1991.
Preliminary phylogenetic systematic analysis of the
major lineages of the Eucestoda (Platyhelminthes:
Cercomeria). Proceedings of the Biological Society of
Washington, 104, pp. 651-688
Chandler, A. C., 1946. Observations on the anatomy of
Mesocestoides. Journal of Parasitology, 32, pp. 242-
246.
Coleman, A.W., 2003. ITS2 is a double-edged tool for
eukaryote evolutionary comparisons. Trends in
Genetics, 19(7), pp.370-375.
Cote, C. A., Peculis, B. A., 2001. Role of the ITS 2-
proximal stem and evidence for indirect recognition of
processing sites in pre-rRNA processing in yeast.
Nucleic Acids Research, 29, pp. 2106-2116.
Dayhoff, M . O., Eck, R. V., Chang, M . A., Sochard, M.
R., 1965. Atlas of Protein Sequence and Structure.
Silver Spring, Maryland: National Biomedical
Research Foundation
Kamegai, S., Ichihara, A., Nonobe, H., Machida, M.,
1967. The 6th and 7th records of human infection
with Mesocestoides lineatus (Cestoda) in Japan.
Research Bulletin of the Meguro Parasitological
Museum, 1, pp. 1-7.
Khalil, L. F., Jones, A., Bray, R. A. eds., 1994. Keys to the
cestode parasites of vertebrates. Wallingford: CAB
International.
Mariaux, J., 1996. Cestode systematics: Any progress?
International Journal for Parasitology, 26(3), pp.
231–243.
Nei, M., Kumar, S., 2000. Molecular Evolution and
Phylogenetics. New York: Oxford University Press.
Page, R.D., 1996. TreeView: an application to display
phylogenetic trees on personal computers. Computer
Applications in the Biosciences, 12, pp. 357-358.
Ronquist, F., Huelsenbeck, J. F., 2003. MRBAYES 3:
Bayesian phylogenetic inference under mixed models.
Bioinformatics,19, pp. 1572-1574.
Saitou, N., Nei, M., 1987. The neighbor-joining method:
A new method for reconstructing phylogenetic trees.
Molecular Biology and Evolution, 4, pp. 406-425.
Schultz, J., Maisel, S., Gerlach, D., Müller, T., Wolf, M.,
2005. A common core of secondary structure of the
internal transcribed spacer 2 (ITS2) throughout the
Eukaryota. RNA, 11(4), pp. 361-364.
Siebert, S., Backofen, R., 2005. MARNA:multiple
alignment and consensus structure prediction of RNAs
based on sequence structure comparisons.
Bioinformatics, 21, pp. 3352-3359
Tamura, K., Dudley, J., Nei, M., Kumar, S., 2007.
MEGA4: Molecular Evolutionary Genetics Analysis
(MEGA) software version 4.0. Molecular Biology &
Evolution, 24 (8), pp.1596-1599.
Tamura, K., Dudley, J., Nei, M., Kumar, S., 2004.
SYSTEMATIC POSITION AND PHYLOGENETIC RELATIONSHIPS OF THE CYCLOPHYLLIDEAN CESTODES -
An In-silico Study using ITS2 rDNA and Sequence-structure Alignment
11

Prospects for inferring very large phylogenies by using
the neighbor-joining method. Proceedings of the
National Academy of Sciences,101, pp.11030-11035.
Thompson, J. D., Higgins, D. G., Gibson, T. J., 1994.
CLUSTAL W: improving the sensitivity of
progressive multiple sequence alignment through
sequence weighting, position-specific gap penalties
and weight matrix choice. Nucleic Acids Research,
(22), pp. 4673-80.
von Nickisch-Rosenegk, M., Lucius, R., Loos-Frank, B.,
1999. Contributions to the phylogeny of the
Cyclophyllidea (Cestoda) inferred from mitochondrial
12S rDNA. Journal of Molecular Evolution, 48(5), pp.
586-596.
Wolf, M., Ruderisch, B., Dandekar, T., Schultz, J., Müller,
T., 2008. ProfDistS: (profile-) distance based
phylogeny on sequence--structure alignments.
Bioinformatics, 24(20), pp.2401-2402.
Zuker, M., 2003. Mfold web server for nucleic folding and
hybridization prediction. Nucleic Acids Research, 31,
pp. 3406-3415.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
12
