
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS
OF PANCREATIC CANCER
Stuart Floyd, Carolina Ruiz
Department of Computer Science, Worcester Polytechnic Institute, 100 Institute Road, Worcester, MA 01609, U.S.A.
Sergio A. Alvarez
Department of Computer Science, Boston College, 140 Commonwealth Avenue, Chestnut Hill, MA 02467, U.S.A.
Jennifer Tseng, Giles Whalen
Department of Surgical Oncology, University of Massachusetts Medical School, Worcester, MA 01605, U.S.A.
Keywords: Machine learning, Meta-learning, Cancer, Prognosis.
Abstract: Machine learning predictive techniques have been shown to be useful in establishing cancer prognosis.
However, no single machine learning technique provides the best results in all cases. This paper introduces
an automated meta-learning technique that learns to predict the best performing machine learning technique
for each patient. The individually selected machine learning technique is then used for prognosis for the
given patient. The performance of the proposed approach is evaluated over a database of retrospective
records of pancreatic cancer resections.
1 INTRODUCTION
Despite progress in the treatment of pancreatic
adenocarcinoma over the past two decades, this
disease remains one of the most lethal of all cancers.
The five year survival rate is less than 6%, based on
the most recent (2009) National Cancer Institute data
for cancers diagnosed between 1999 and 2005
(SEER, 2009). Nonetheless, there are groups of
patients for which the outlook is significantly better.
Cancer stage at diagnosis is of particular importance.
For example, the survival rate for localized cancers
is fully four times the average. The results of
specific diagnostic tests and individual patient
attributes including age also affect prognosis.
1.1 Machine Learning for Cancer
Prognosis
Machine learning refers to a set of techniques,
including decision tree induction, neural and
Bayesian network learning, and support-vector
machines, in which a predictive model is constructed
or “learned” from data in a semi-automated fashion
(e.g., Mitchell, 1997). In supervised learning, which
is the sort considered in the present paper, each data
instance used for learning (training) consists of two
portions: an unlabeled portion, and a categorical or
numerical label known as the class or target attribute
that is provided by human experts. The object of
learning is to predict each data instance’s label based
on the instance’s unlabeled portion. The result of
learning is a model that can be used to make such
predictions for new, unlabeled data instances.
Machine learning has been successfully applied
to pancreatic cancer detection (Honda et al., 2005)
and to the analysis of proteomics data in pancreatic
cancer (Ge and Wong, 2008). Machine learning
techniques have also been shown to provide
improved prediction of pancreatic cancer patient
survival and quality of life when used either instead
of, or together with, the traditional technique of
logistic regression (Floyd et al., 2007; Hayward et
al., 2008).
The quality of the predictions produced by a
given machine learning method varies across
29
Floyd S., Ruiz C., A. Alvarez S., Tseng J. and Whalen G. (2010).
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS OF PANCREATIC CANCER.
In Proceedings of the Third International Conference on Health Informatics, pages 29-37
DOI: 10.5220/0002711000290037
Copyright
c
SciTePress
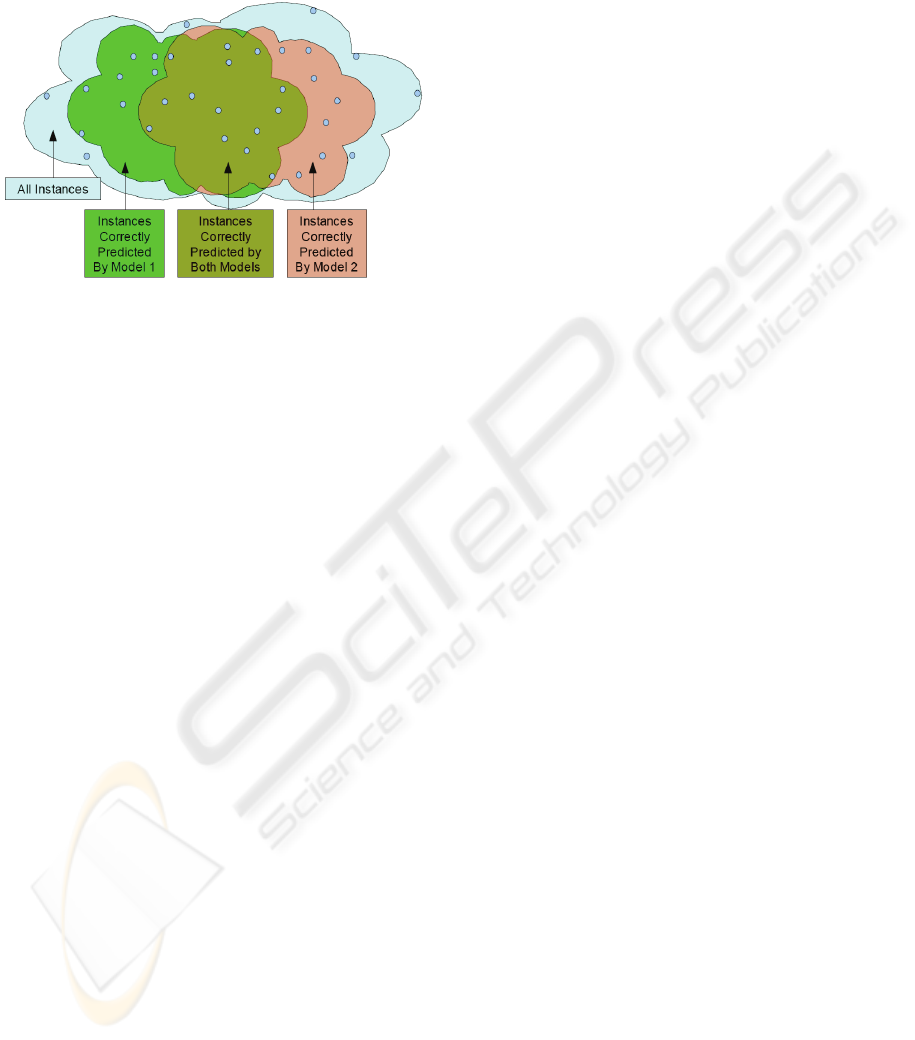
patients. In particular, the method that provides the
best predictive model for one patient will not
necessarily be optimal for another patient. An
example is suggested pictorially in Figure 1.
Figure 1: Sets of instances classified correctly by different
models.
The latter fact suggests that overall predictive
performance across all patients could be improved if
it were possible to reliably predict, for each patient,
what machine learning method will provide the best
performance for that particular patient. The selected
method can then be used to make predictions for the
patient in question. This is the basic idea behind the
approach described in the present paper.
1.2 Classical Meta-learning
Several “meta-learning” approaches have been
developed in machine learning, including bagging,
boosting, and stacking (see below). These
approaches are also known as ensemble methods
because they aggregate the predictions of a
collection of machine learning models to construct
the final predictive model. Ensemble machine
learning methods have previously been applied to
cancer (Qu et al., 2002; Bhanot et al., 2006; Ge and
Wong, 2008). The present paper describes a new
ensemble machine learning approach and its
application to prognosis in pancreatic cancer.
In bagging (Breiman, 1996), the models in the
ensemble are typically derived by applying the same
machine learning technique (e.g., decision tree
induction, or neural network learning) to several
different random samples of the dataset over which
learning is to take place. The bagging prediction is
made by a plurality vote taken among the learned
models in the case of categorical classification, and
by averaging the models’ predictions in the case of a
numerical target. In boosting (Freund and Schapire,
1997), a sequence of models is learned, usually by
the same learning technique, with each model
focusing on data instances that are poorly handled
by previous models. The overall boosting prediction
is made by weighted voting among the learned
models. Stacking (Wolpert, 1992) allows the use of
different machine learning techniques to construct
the models over which aggregation is to take place.
In this context, the individual models are known as
level 0 models. The outputs of the level 0 models are
then viewed as inputs to a second layer of learning,
known as the level 1 model, the output of which is
used for prediction.
1.3 Proposed Model Selection
Meta-learning
The model selection approach proposed in the
present paper is an ensemble meta-learning approach
in that it involves learning a collection of models.
Our approach is more similar to boosting and
stacking than to bagging in its use of the full training
dataset to learn the individual models. However, it
differs from classical bagging, boosting, and
stacking, and is characterized by, its adoption of a
new prediction target. Rather than aiming to predict
the original target, say survival, directly, the goal
changes in our approach to identifying what learned
model is best qualified to make the desired
prediction for a given data instance. Once identified,
the selected model alone is used to predict the
original target.
1.4 Plan of the Paper
Section 2 describes the pancreatic cancer patient
database that was constructed for our work. Section
3 presents the details of the model selection meta-
learning method proposed in the present paper.
Section 4 describes the results of an experimental
evaluation of model selection meta-learning over
pancreatic cancer data.
2 PANCREATIC CANCER
DATASETS
A clinical database was assembled containing
retrospective records of 60 patients treated by
resection for pancreatic adenocarcinoma at the
University of Massachusetts Memorial Hospital in
Worcester. Each patient record is described by 190
fields comprising information about preliminary
outlook, personal and family medical history,
HEALTHINF 2010 - International Conference on Health Informatics
30
diagnostic tests, tumor pathology, treatment course,
surgical proceedings, and length of survival.
A summary of the categories of attributes and the
number of attributes in each category is presented in
the Table 1. Note that the attributes are divided into
three major categories: 111 pre-operative attributes,
78 peri-operative attributes, and the target attribute.
Table 1: Attribute categories for the pancreatic cancer
database.
Category Number of
attributes
Category
Description
Pre-operative attributes
Patient 6 Bio. info. patient
Presentation 21 Symptoms at
diagnosis
History 27 Past health history
Serum 8 Lab test scores
Diagnostic
imaging
23 Imaging scan details
Endoscopy 25 Endoscopy details
Prelim. Outlook 1 Physician’s pre-
surgical evaluation
Total 111
Peri-operative attributes
Treatment 36 Treatment details
Resection 24 Surgical removal
details
Pathology 7 Post-surgical tumor
type results
No Resection 11 Reasons for tumor
non-removal
Total 78
Target attribute
Survival 1 Time between
diagnosis and death
Grand Total 190
The prediction target (or target attribute) of
our analysis is survival time, measured as the
number of months between diagnosis and death. In
this work, we considered different binnings of this
target attribute:
• 9 month split, resulting in 2 target values: <9
months (containing 30 patients), and >9 months
(30 patients).
• 6 month split, resulting in 2 target values: <6
months (20 patients), and >6 months (40
patients).
• 6 and 12 month splits, resulting in 3 target
values: less than 6 months (20 patients), 6 to 12
months (20 patients), and over 12 months (20
patients).
Also, we consider two subsets of attributes of this
dataset: one containing all 190 attributes (denoted by
“All-Attributes Dataset”), and one containing only
the 111 pre-operative attributes together with the
target attribute (denoted by “Pre-Operative
Dataset”). In our experimentation we consider a total
of 6 datasets determined by the 2 subset of attributes
used and the 3 types of binning of the target
attribute.
3 OUR MODEL SELECTION
META-LEARNING
TECHNIQUE
The present paper proposes a new meta-learning
approach based on predicting for each data instance
the machine learning model that is best suited to
handle that instance. We refer to this approach as
model selection meta-learning. The motivation
behind our approach is visually depicted in Figure
1, in which each of two models correctly covers only
a subset of the instances. If we could correctly
predict which model to use for each instance, overall
classification performance would be improved.
3.1 Model Selection Meta-learner for
Prediction
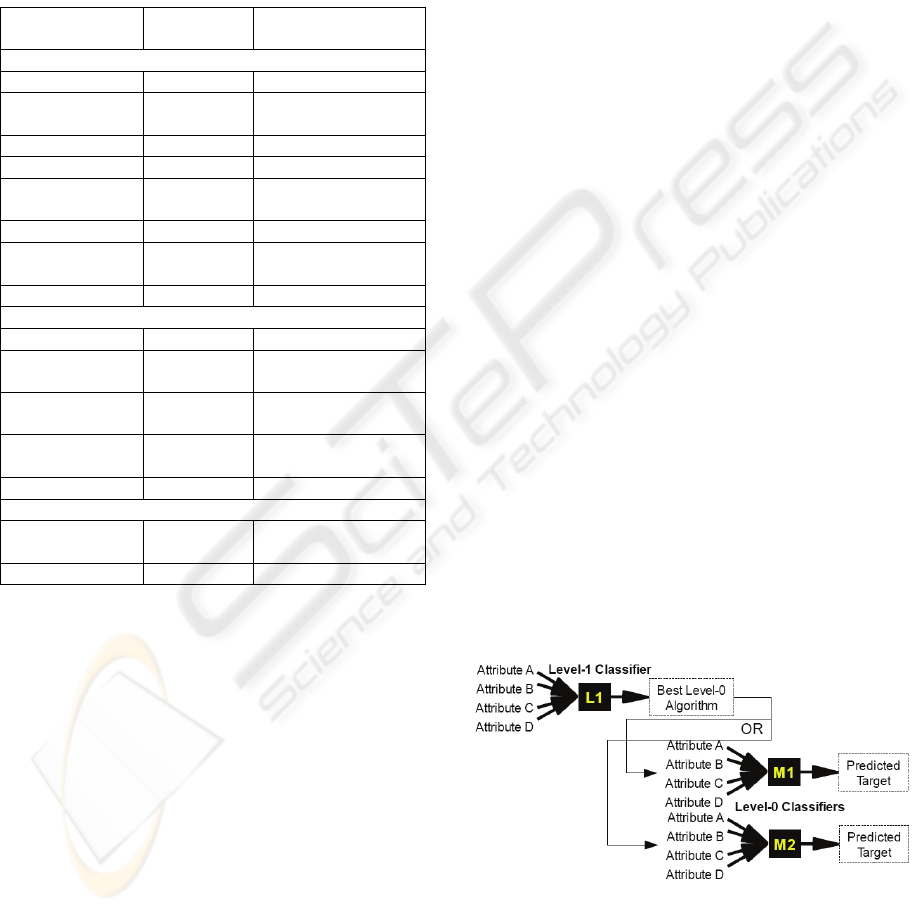
Figure 2 shows how the proposed model selection
meta-learner uses two levels of classifiers to predict
the unknown target class of a set of a previously
unseen input instance. After the level 1 classifier
predicts which of the level 0 models is expected to
perform best on the given instance, the instance’s
attributes are run through the selected model to
make a prediction.
Figure 2: Model selection meta-learner.
The prediction process is described in
pseudocode below. It is assumed here that the meta-
learner has previously been trained (see Section 3.3).
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS OF PANCREATIC CANCER
31
For a previously unseen data instance:
1. Run data instance through level 1 classifier to
select which level 0 classifier to use. We
assume that the learned level 0 models
produce class probability distributions
(p
1
…p
k
) as their outputs for a given input
instance x, with each p
j
an estimate of the
conditional probability P(target value j | x)
that the given instance has target value j
(e.g., the conditional probability of a patient
surviving > 6 months given the patient’s
data). See Figure 3 for an example in which
the target attribute is binary with possible
values “+” and “-”. These numerical
outputs provide the basis for the selection
of a “best” model during meta-learning. In
brief, the “best” model is the one that
outputs the highest posterior probability
P(correct target value for x | x) for the input
instance x.
2. Run data instance through level 0 classifier
recommended by level 1 classifier to
predict the target value of the instance (e.g.,
survival time of the patient).
Our model selector meta-learning approach is
similar to stacking in that it uses a collection of level
0 machine learning models followed by a level 1
learner. The key difference is in the function of the
level 1 meta-learner. Stacking's level 1 classifier
combines the target class probability distributions
generated by running the unseen instance through
each of the level 0 models, while our model
selector's level 1 classifier selects which of the level
0 models is expected to output the highest
probability for the correct class of the given test
instance. Despite this fundamental difference with
stacking, we will use the level 0 and level 1 stacking
terminology throughout, for convenience.
3.2 Training the Level 0 Models
A level 0 model is obtained by applying a machine
learning technique to the input dataset. We will
denote that dataset by I
0
. As explained in Section
3.1, we assume that the prediction that the trained
model outputs is a probability distribution over the
possible target values. In our case, the input dataset
is the pancreatic cancer dataset described in Section
2. Hence each level 0 model is trained to predict the
survival time of patients. The prediction output by
the trained model is then a probability distribution
over the possible survival time values. For instance,
if the 6 and 12 month splits are used, then given a
patient’s data, the trained level 0 model will output
the probabilities that the patient will survive < 6
months, between 6 and 12 months, and > 12 months.
3.3 Training the Level 1 Model
In section 3.1 we described how the model selection
meta-learner is used to predict the target class of a
new instance, assuming that the meta-learner has
previously been trained. We now describe how the
training is carried out. A two-stage approach is used.
First, a new dataset I
1
is constructed from the
original dataset of instances I
0
using cross
validation, by relabeling each training instance with
the name of the level 0 model that outputs the
highest probability for that instance’s correct target
class; this model is considered to be the best
predictor for the given instance. In the second stage,
the level 1 model is trained over the new dataset I
1
.
Once the level 1 model has been trained, level 0
models are retrained over the full original dataset as
described in section 3.2.
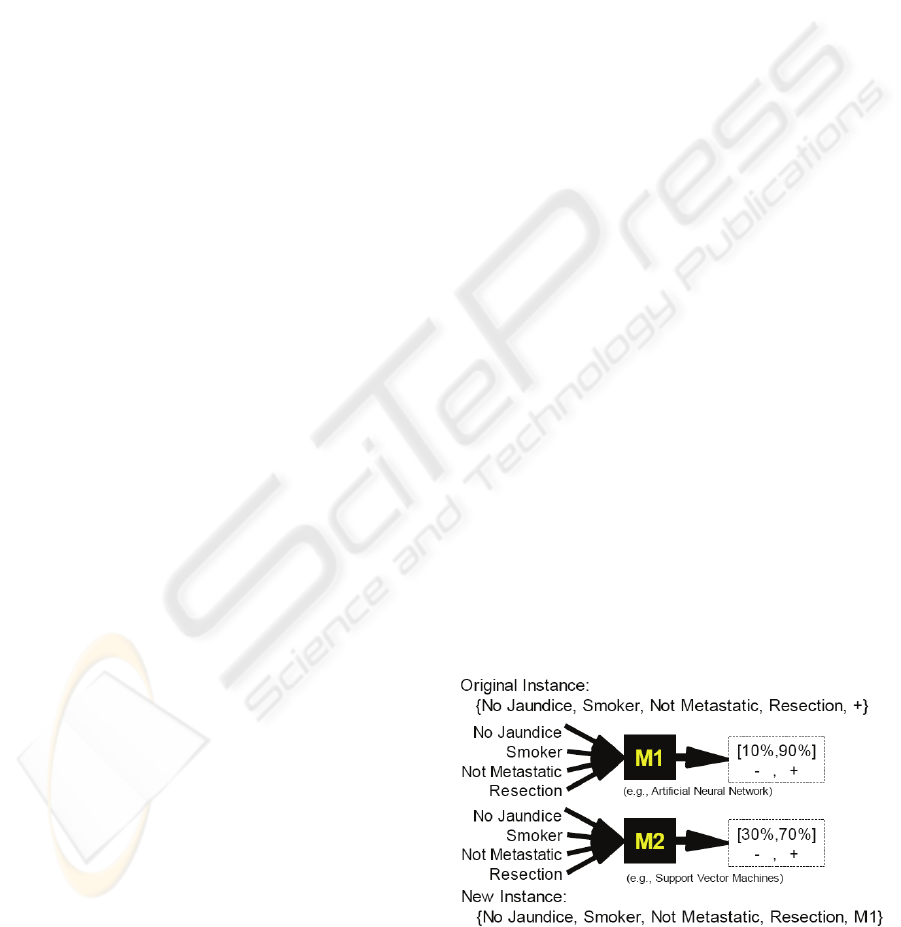
Example. An example to illustrate the construction
of the dataset to train the level 1 model is shown in
Figure 3. The example patient in the figure with the
given medical history as the input attributes is
known to fall into the positive class (say survival
time > 6 months). When M1 uses this set of
symptoms to predict that there is a 90% chance that
this patient is in the positive class and M2 predicts
that there is a 70% chance that this patient is in the
positive class. Since M1 has the highest confidence
in the correct classification, this is the model said to
best predict this instance. A new instance is then
created using the patient's medical history as the
input attributes and M1 as the target value. This
new instance is added to the dataset used to train the
level 1 model.
Figure 3: Transformation of a level 0 data instance into a
level 1 instance.
HEALTHINF 2010 - International Conference on Health Informatics
32

The model selector meta-learning algorithm is
described in greater detail below.
Training Algorithm:
Inputs:
- Set I
0
of input data instances
(in our case, each input instance corresponds to
the data of a pancreatic cancer patient labeled
with the patient’s survival time as a nominal
range)
- Set of level 0 machine learning techniques to
use, each of which outputs a class probability
distribution
- Level 1 machine learning technique to use
- Integer n (user selected number of folds in
which to split the input dataset)
Output: Trained Level 1 model
(A) Construct a new dataset of training instances I
1
(to be used to train the level 1 classifier) as follows:
1. Initialize I
1
as empty
2. Randomly split I
0
into n stratified folds: I
0
1
,…,
I
0
n
.
3. For each fold I
0
i
, with 1 ≤ i ≤ n, and for each
level 0 machine learning technique received as
input:
a. Train a level 0 classifier using the machine
learning technique and the data instances in
the union of all the n folds except for fold
I
0
i
b. For each data instance d in fold I
0
i
:
o Run each level 0 classifier on d. Each
will output a probability distribution of
the target values.
o Among the level 0 classifiers, select
one with the highest probability for d's
correct target value. This correct target
value is given in the input training data
I
0
.
o Add instance d to I
1
replacing its
original target value with the identifier
of the level 0 classifier selected above
(e.g., if d corresponds to a patient
whose survival time was more than 12
months, and the neural network was
the classifier with the highest
probability for “more than 12 months”,
then the instance d will appear in I
1
with its target value (“more than 12
months”) replaced by “neural
network”.
(B) Train the level 1 classifier using the dataset I
1
.
(C) Rebuild each level 0 classifier over all training
instances in I
0
.
4 EVALUATION OVER
PANCREATIC CANCER DATA
We discuss in this section the experimental
evaluation that we performed over the database of
pancreatic cancer resections that we constructed,
which is described Section 2.
4.1 Data Mining Techniques used
Feature Selection. We use Attribute Selection to
evaluate models built with different machine
learning algorithms using the features selected by
various feature selection algorithms. Previous work
in pancreatic cancer (Ge and Wong, 2008; Hayward
et al., 2008) has shown that feature selection can
improve the prediction performance of classification
methods. In the current paper we investigate the use
of the Gain Ratio, Principal Components Analysis
(PCA), ReliefF, and Support Vector Machines
(SVMs) for feature selection. All of these algorithms
rank order the most important features, allowing the
number of features retained to be prescribed.
Through these experiments we attempt to determine
the optimal feature selection approach for a given
machine learning algorithm.
Machine Learning Techniques for Level 0 and
Level 1 Classifiers. We consider artificial neural
networks (ANNs), Bayesian networks (BNs),
decision trees, naïve Bayes networks, and support
vector machines (SVM). The first three classification
methods above have previously been identified
(Hayward et al., 2008) as the most accurate over a
pancreatic cancer dataset among a wide range of
methods. SVM is included in the present work both
as a feature selection method and as a classification
method.
For each dataset, we find the best combination of
feature selection and machine learning algorithm.
We use ZeroR (majority class classifier) and logistic
regression as benchmarks against which to compare
the performance of the models constructed.
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS OF PANCREATIC CANCER
33
4.2 Experimental Protocol
All experiments reported here were carried out using
the Weka machine learning toolkit (Witten and
Frank, 2005). The classification accuracy for all
experiments is calculated as the average value over
ten repetitions of 10-fold cross validation, each
repetition with a different initial random seed (for a
total of 100 runs of each experiment).
For each of the 6 datasets described in Section 2,
we apply the following procedure systematically:
1. Select the Level 0 Classifiers. We applied each
of the machine learning techniques under
consideration with and without feature selection to
the dataset and recorded the resulting accuracy
reported by the 10 repetitions of 10-fold cross
validation procedure described above. For each of
the machine learning techniques, all the feature
selection approaches were tested with a varying
number of attributes to be selected. In most cases,
feature selection increased the accuracy of the
machine learning methods. Then we selected the top
3 most accurate models among all models: the ones
with and the ones without feature selection.
2. Select the Level 1 Classifier. Once the top 3
performing level 0 models were identified, we ran
experiments to determine what subset of those 3 top
models together with what level 1 machine learning
technique would yield the model-selector meta-
classifier with the highest predictive accuracy. As
above, all machine learning techniques with and
without feature selection (and allowing the size of
the selected attribute set to vary) were considered.
Note than in this case, feature selection is applied to
the level 1 dataset, not to the original dataset. The
model selector meta-classifier with highest
predictive accuracy is reported.
4.3 Results and Discussion
We describe the results of our experimental
evaluation, focusing on the pre-operative dataset
described by 111 attributes.
4.3.1 Pre-operative Dataset, 9 Month Split
• Individual Machine Learning Methods. The
classification accuracies obtained by individual
machine learning methods with no attribute selection
appear in Table 2. The ZeroR, or majority class,
classifier in the first row is a simple benchmark that
selects the most frequent class for all instances. For
a 9-month split, the two classes occur equally
frequently in the dataset; the ZeroR prediction
amounts to a random choice between them.
• Feature Selection. Attribute selection allowed
classification performance to be improved slightly.
The best results were obtained using GainRatio
attribute selection in conjunction with either a
logistic regression classifier (65.5% accuracy) or an
SVM classifier (65.5% accuracy), and ReliefF
attribute selection with a Bayesian network classifier
(65.3% accuracy).
Table 2: No Feature Selection, Nine Month Split.
Machine Learning Algorithm
Classification
Accuracy %
ZeroR 50.0
Logistic Regression 58.8
SVM 62.5
ANN 58.5
Naïve Bayes 49.3
C4.5 decision tree 49.5
Bayesian Network 64.7
• Comparison of Model Selection Meta-learning
with other Techniques. Table 3 shows the
classification accuracies of the best performing
classifier / feature selection combinations for 9
month split, together with the accuracy of the model
selection meta-learning approach proposed in the
present paper. The proposed approach slightly
outperforms the best individual level 0 classifier
methods. In passing, we note that the model
selection meta-classifier also outperformed the
standard meta-learning techniques of bagging,
boosting, and stacking.
Table 3: Classification accuracy: pre-operative attributes
only, nine month split.
Machine Learning (ML)
Technique(s)
Feature
Selection
(FL)
#
attrib
utes
Accu
racy
%
Pre-Operative Attributes only, 9 month split
Best Performing ML + FL Combinations:
Logistic Regression GainRati
o
70 65.5
SVM GainRati
o
80 65.5
Bayesian Net ReliefF 100 65.3
Best Performing Model Selection Meta-Classifier:
Level 1: Naïve Bayes
Level 0: Logistic, SVM
None
GainRati
o
111 67.3
HEALTHINF 2010 - International Conference on Health Informatics
34
4.3.2 Pre-operative Dataset, 6 Month Split
Since this dataset contains 20 patients with survival
time < 6 months and 40 patients with > 6 months,
the accuracy of ZeroR (majority class) classification
is 66.6%. Table 4 shows the top three combinations
of machine learning classification and feature
selection obtained, and the most accurate level 1
classifier constructed over them. Once again our
model selection meta-learning method outperformed
bagging, boosting, and stacking.
Table 4: Classification accuracy: pre-operative attributes
only, six month split.
Machine Learning (ML)
Technique(s)
Feature
Selection
(FL)
#
attrib
utes
Accu
racy
%
Pre-Operative Attributes only, 6 month split
Best Performing ML + FL Combinations:
Logistic Regression GainRati
o
40 70.2
SVM GainRati
o
30 69.8
ANN GainRati
o
30 68.5
Best Performing Model Selection Meta-Classifier:
Level 1: Logist.
Regression
Level 0: Log. Regr.,
SVM
PCA
GainRati
o
15 70.8
4.3.3 Pre-operative Dataset with 6 and 12
Month Splits
Classification performance results for the three class
dataset obtained by splitting the target attribute at
both 6 and 12 months appear in Table 5. The
accuracy values are lower than in Table 3 and Table
4 because of the larger number of classes (3 vs. 2).
For comparison, randomly guessing the class would
lead to an accuracy of approximately 33.3% for this
dataset, as the three classes are equally frequent.
Model selection meta-learning once again slightly
outperforms the individual level 0 models.
4.3.4 All-attributes Dataset with 6 Month
Split
We discuss here the best meta-classifier obtained via
the approach presented in this paper for the All-
Attributes dataset with 6 months split, as it
illustrates several interesting points. The
classification accuracy of this model (75.2%) is
significantly greater than that of logistic regression
(61.3%), the most widely accepted statistical method
Table 5: Classification accuracy: pre-operative attributes
only, 6 and 12 month splits.
Machine Learning (ML)
Technique(s)
Feature
Selection
(FL)
#
attri
bute
s
Accu
racy
%
Pre-Operative Attributes only, 6 and 12 month splits
Best Performing ML + FL Combinations:
Bayesian Net ReliefF 20 52.7
ANN GainRatio 50 51.8
SVM ReliefF 20 48.5
Best Performing Model Selection Meta-Classifier:
Level 1: Naïve Bayes
Level 0: ANN
SVM
None
GainRatio
,
ReliefF
111 53.3
in the medical community. Meta-classifier accuracy
also exceeds that of majority classification (66.6%).
This meta-classifier constructed by our model
selector combines the models constructed by two top
performing level 0 classifiers: Naïve Bayes (using
Gain Ratio feature selection) and Artificial Neural
Network (using Gain Ratio feature selection also). A
C4.5 decision tree (J4.8 in Weka) coupled with
SVM feature selection was used as the level 1
classifier. As in our other experiments, the resulting
model selection meta-classifier was superior in
prediction performance (75.2% accuracy) to the best
models constructed with the standard meta-learning
techniques of bagging (74.5%), boosting (67%), and
stacking (72.5%).
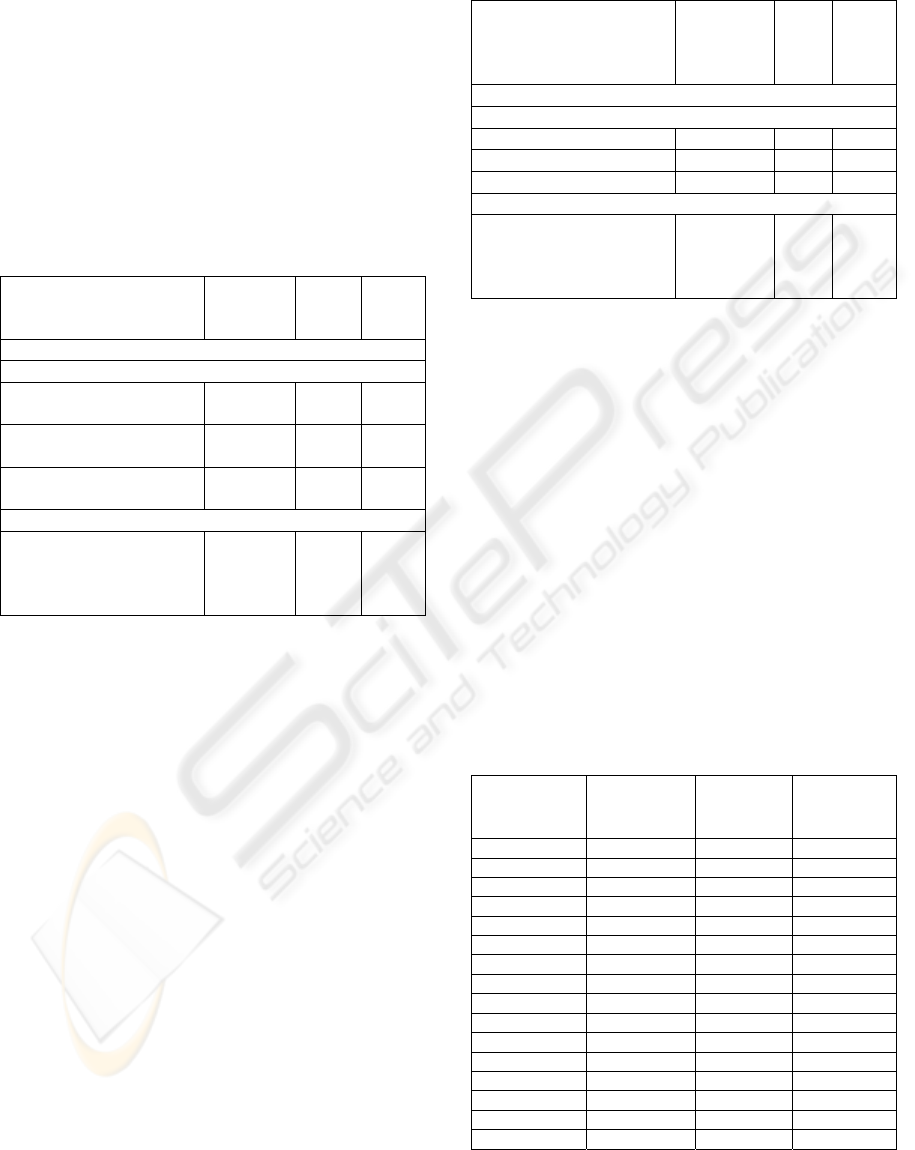
Table 6: Class probability distributions and correct level 0
models for a subset of data instances. {x,y} values are
probabilities of survival for less than 6 months (x) and at
least 6 months (y).
ANN class
probabilities
Naïve
Bayes class
probs.
Actual
Target
Value
Correct
Level 0
Model(s)
{0.72,0.28} {0.12,088} < 6 months ANN
{0.88,0.12} {0.39,0.61} < 6 months ANN
{0,1} {0.85,0.15} < 6 months Naïve Bayes
{0.95,0.05} {0.08,0.92} > 6 months Naïve Bayes
{0.99,0.01} {0.48,0.52} > 6 months Naïve Bayes
{0.59,0.41} {0.47,0.53} > 6 months Naïve Bayes
{0.99,0.01} {0.85,0.15} < 6 months Both
{0.05,0.95} {0.09,0.91} > 6 months Both
{0,1} {0.39,0.61} > 6 months Both
{0.01,0.99} {0.45,0.55} > 6 months Both
{0.07,0.93} {0.1,09} > 6 months Both
{0.01,0.99} {0.12,0.88} > 6 months Both
{0,1} {0.1,0.9} > 6 months Both
{0.1} {0.1,0.9} < 6 months Neither
{0.03,0.97} {0.11,0.89} < 6 months Neither
{0.06.0.94} {0.07,0.93} < 6 months Neither
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS OF PANCREATIC CANCER
35

Table 6 shows the class probability distributions
for selected instances over each of the two level 0
models. The actual target value is also in the table
along with a label stating which of the two models
(or both) predict this value correctly, or if neither of
the models predicts the target value correctly. In 36
out of these 60 instances both models produce the
correct classification (31 of which are > 6 months,
and 5 are < 6 months). In eight instances neither
model produces the correct prediction (which is < 6
months for all eight instances). This leaves 16
instances for which picking the right model would
lead to making the correct prediction: in 11 of these
naïve Bayes is correct (of which 2 are < 6 months),
and in 5 ANN is correct (of which all are < 6
months). An interesting observation is that when the
artificial neural network and the naïve Bayes model
both predict the same target, the artificial neural
network is much more certain of its prediction.
As mentioned above, SVM feature selection was
applied to the level 1 training dataset reducing the
number of attributes from 190 to 70. Remarkably all
these 70 selected attributes are pre-operative. We
describe below this resulting set of 70 attributes by
grouping them into related categories:
Presentation - Demographic (3 attributes selected):
Patient's Height, Weight, and Quality-of-life score
(ECOG) at admission
Presentation - Initial Symptoms (18 attributes
selected): Abdominal pain, Back pain, Biliary colic,
Clay colored stool, Cholecystitis, Cholangitis,
Dysphagia, Fatigue, Indigestion, Jaundice, Nausea,
Pruritis, Early Satiety, Vomiting, and Weight Loss.
Presentation - Presumptive Diagnosis (1 attribute
selected): Initial diagnosis (e.g., pancreatic tumor,
periampullary tumor, etc.).
Medical History - Comorbidities (3 attributes
selected): Heart Failure, Ischemic Heart Disease,
and Respiratory Diseases.
Serum Laboratory Tests (8 attributes selected):
Albumin, Alkaline phosphotase, ALT (alanine
transaminase), AST (aspartate aminotransferase),
Bilirubin, Amylase, CA19-9 (carbohydrate antigen
19-9), and CEA (carcinoembryonic antigen).
Diagnostic Imaging - Computer Tomography
(CT) (19 attributes selected): Celiac Artery
Involvement, Celiac Nodal Disease, Hepatic Vein
Involvement, Inferior Vena Cava Involvement,
Lymph node disease or other nodal disease, Node
Omission, Portal Vein Involvement, Superior
Mesenteric Artery Involvement, Superior Mesenteric
Vein Involvement, Tumor Height (cm), Tumor
Width (cm), Vascular Omission, and CT Diagnosis.
Diagnostic Imaging - Endoscopic UltraSound
(EUS) (15 attributes selected): Virtually the same
attributes as for CT, and EUS Diagnosis.
Diagnostic Imaging - Chest X-Rays (1 attribute):
Chest X-Ray Diagnosis.
Diagnostic Imaging - Percutaneous Transhepatic
Cholangiography (PTC) (3 attributes selected): If
stent was used and what type, and PTC diagnosis.
The level 1 machine learning technique used here is
C4.5 decision trees (J4.8 in Weka). The resulting
pruned decision tree is included below. Out of the 70
attributes, only 6 are used in the pruned decision
tree: 2 initial symptoms (presentation), including
back pain (which was shown to be an important
attribute by the Bayesian Nets constructed in other
of our experiments) and the occurrence of jaundice;
2 results of diagnostics imaging tests (CT and EUS);
and 2 serum lab tests (Bilirubin and Albumin).
If patient presents Back Pain
| if CT shows Node Omission
| then Use Naïve Bayes
| else
| | if Bilirubin Serum Lab Test ≤ 0.9
| | then Use Naïve Bayes
| | else Use Artificial Neural Net
else (* patient does not present Back Pain *)
| if patient presents Jaundice
| then
| | if EUS shows Vascular Omission
| | then Use Naïve Bayes
| | else
| | | if Albumin Serum Lab Test ≤ 2.4
| | | then Use Naïve Bayes
| | | else Use Artificial Neural Net
| else Use Artificial Neural Net
5 CONCLUSIONS AND FUTURE
WORK
This paper has presented a new approach to
combination of machine learning methods through
meta-learning, and an evaluation of this technique
for pancreatic cancer prognosis using a database of
retrospective patient records. The proposed
technique, model selection meta-learning, is based
on learning which of several available machine
learning methods can be expected to be the best
predictor for a given input instance. The motivation
HEALTHINF 2010 - International Conference on Health Informatics
36

for this technique is the fact that different methods
sometimes produce conflicting predictions for the
same instance. Thus, a system that reliably identifies
the best predictor for a given instance will achieve
better predictive performance than any of the
individual predictors. The experimental evaluation
presented in this paper focuses on predicting
survival time of pancreatic cancer patients based on
attributes such as demographic information, initial
symptoms, and diagnostic test results. The
evaluation results show that the proposed technique
of model selection meta-learning produces
predictions that are better than those of the
individual machine learning methods. Also, the
proposed technique outperforms the standard meta-
learning techniques of bagging, boosting, and
stacking in the experiments conducted for this paper.
Further work is needed to better establish the
magnitude of observed performance differences, and
to determine whether any particular machine
learning predictors are best suited to being combined
through the model selection meta-learning technique
introduced in this paper.
REFERENCES
Bhanot, G., Alexe, G., Venkataraghavan, B., Levine, A.J.
A robust meta-classification strategy for cancer
detection from MS data, Proteomics 2006, 6:592-604.
Breiman, L.. Bagging predictors, Machine Learning 24(2):
123-140, 1996.
Floyd, S., Alvarez, S. A., Ruiz, C., Hayward, J., Sullivan,
M., Tseng, J., and Whalen, G. Improved survival
prediction for pancreatic cancer using machine
learning and regression, Society for the Surgery of the
Alimentary Tract 48th Annual Meeting (SSAT 2007),
Washington DC, USA, May 19-23, 2007.
Freund, Y. and Schapire, R.E. A decision-theoretic
generalization of on-line learning and an application to
boosting, Journal of Computer and System Sciences,
55(1):119--139, 1997.
Ge, G. and Wong, G.W. Classification of premalignant
pancreatic cancer mass-spectrometry data using
decision tree ensembles, BMC Bioinformatics 2008,
9:275
Hayward, J., Alvarez, S.A., Ruiz, C., Sullivan, M., Tseng,
J., and Whalen, G. Knowledge discovery in clinical
performance of cancer patients, IEEE International
Conference on Bioinformatics and Biomedicine
(BIBM08), Philadelphia, PA, USA, Nov. 3-5, 2008.
Honda, K., Hayashida, Y., Umaki, T., Okusaka, T.,
Kosuge, T., Kikuchi, S., Endo, M., Tsuchida, A.,
Aoki, T., Itoi, T., Moriyasu, F., Hirohashi, S.,
Yamada, T. Possible detection of pancreatic cancer by
plasma protein profiling. Cancer Res. 2005 Nov 15;
65(22):10613-22.
Horner, M.J., Ries, L.A.G., Krapcho, M., Neyman, N.,
Aminou, R., Howlader, N., Altekruse, S.F., Feuer,
E.J., Huang, L., Mariotto, A., Miller, B.A., Lewis,
D.R., Eisner, M.P., Stinchcomb, D.G., Edwards, B.K.
(eds). SEER Cancer Statistics Review, 1975-2006,
National Cancer Institute. Bethesda, MD,
http://seer.cancer.gov/csr/1975_2006/, based on
November 2008 SEER data submission, posted to
SEER web site, 2009.
Mitchell, T. Machine Learning, McGraw-Hill, 1997.
Qu, Y., Adam, B.L., Yasui, Y., Ward, M.D., Cazares,
L.H., Schellhammer, P.F., Feng, Z., Semmes, O.J.,
Wright, G.L. Jr.: Boosted decision tree analysis of
surface-enhanced laser desorption/ionization mass
spectral serum profiles discriminates prostate cancer
from noncancer patients. Clin Chem 2002, 48:1835-
1843.
Witten, I.H and Frank, E. Data Mining. 2
nd
ed. Morgan
Kaufmann Publishers. 2005.
Wolpert, D.H. Stacked generalization, Neural Networks,
Vol. 5, pp 241-259, 1992.
MODEL SELECTION META-LEARNING FOR THE PROGNOSIS OF PANCREATIC CANCER
37
