
INFORMATION GAIN OF STRUCTURED MEDICAL
DIAGNOSTIC TESTS
Integration of Bayesian Networks and Ontologies
Marin Prcela, Dragan Gamberger, Tomislav Šmuc
Rudjer Boskovic Institute, Bijenicka 54, Croatia
Nikola Bogunović
Faculty of Electrical Engineering and Computing, University of Zagreb, Unska 3, Zagreb, Croatia
Keywords: Knowledge representation, Ontologies, Bayesian networks, Integration, Information gain, Decision support
system, Expert system.
Abstract: Usage of Bayesian networks in medical decision support system is in general case twofold: (1) for obtaining
probabilities of occurrence of medical events (i.e. possible diagnosis) and (2) for obtaining information gain
of actions that can be taken (i.e. diagnostic tests). On the other hand, typical role of ontology is to provide a
framework for definition of medical concepts, their structure and relations among them. In medical practice
diagnostic tests are commonly comprised of number of measurements or sub-tests – a structure which is
straightforwardly described by ontological language. In this paper we are analyzing the information gain of
such structured medical diagnostic tests. The purpose of this analysis is to allow finding (1) which
structured medical diagnostic test is at the given point the most informative one and (2) which elementary
measurements within a given diagnostic test are the most informative ones. Furthermore, we are analyzing
some computational issues which arise in the reasoning process.
1 INTRODUCTION
Bayesian networks (BN) have already demonstrated
their practical value in medical decision support
systems. The most exploited features of such system
are (1) finding probabilities of possible events in the
system (usually probabilities of diagnosis) and (2)
finding information gain (IG) of possible actions
that can be taken (usually medical diagnostic tests).
On the other hand, ontologies have become de facto
standard in medical decision support systems for
formalization of descriptive medical knowledge:
defining domain concepts and relations among them.
Medical domain is particularly suitable for usage
of IG as a decision support parameter. For example,
(Jagt 2002) uses BN to describe probabilistic
relations among medical concepts and uses IG to
find the most informative medical measurement
(test) considering possible final diagnoses (diseases).
In practice, medical diagnostic tests are commonly
comprised of more than one diagnostic parameters,
e.g. laboratory analysis of blood sample measures
levels of glucose, creatinine, cholesterol, urea, etc. It
would be useful to allow the decision support system
to perceive such structured medical test as a
conceptual unit. Still, one should be aware that it is
not necessary to measure all existing parameters
within test; one can choose which parameters are
currently interesting and disregard the others and
thus presumably cut the expenses of medical test and
save some time.
In this paper we are proposing approach which
uses BN for description of probabilistic relations
among medical concepts and measures IG of
composite medical diagnostic tests defined within
ontology. As we will demonstrate, the integration of
those two knowledge formalisms brings in some
additional features for the decision making but also
rises some computational issues in the reasoning
process.
It should also be noted that in medical practice
very often some other (non-medical) factors must be
taken into account: e.g. price of the test, availability
235
Prcela M., Gamberger D., Šmuc T. and Bogunovi
´
c N. (2010).
INFORMATION GAIN OF STRUCTURED MEDICAL DIAGNOSTIC TESTS - Integration of Bayesian Networks and Ontologies.
In Proceedings of the Third International Conference on Health Informatics, pages 235-240
DOI: 10.5220/0002713902350240
Copyright
c
SciTePress
of medical instruments, urgency, etc. Although
measure of IG does not take into account these
factors, it is possible to derive a weighted
combination (or some other type) of those factors to
form a comprehensive scale of medical test utility.
However this paper analyzes solely IG of actions in
pure medical sense.
The organization of paper is as follows. Chapter
2 previews the existing approaches for integration of
ontologies and BN. Chapter 3 demonstrates the
usage of BN in a medical decision support system.
Chapter 4 upgrades the described decision support
services with ontological knowledge and analyzes
means for performing reasoning tasks. Chapter 5
gives example of practical usage of such integrated
knowledge base in a single decision support system.
Chapter 6 discusses some performance issues of the
described approach related to reasoning phase.
2 RELATED WORK
Integration of ontologies with BN is not a new
paradigm. In (Pan 2005) BN is used to recognize
semantic relations between concepts in two different
ontologies, which enables automatic generation of
mappings between ontology concepts. Application
of this approach is described in the domain of
Semantic Web where problem of semantic relation
between ontologies is emphasized.
In (Devitt 2006) knowledge stored in the
ontology is used for generating possible structures of
BN. Since ontologies thoroughly define domain
concepts and existing relations among them there is
a possibility to use such knowledge to generate the
structure of BN. In (Town 2004) ontology is used
both to learn BN structure and in the process of
network training, i.e. learning probability tables of
network nodes. The usual process of BN training
(using existing data set) is augmented by scoring
scheme which is based on the ontological
knowledge.
In (McGarry 2007) high level knowledge
obtained from ontologies is integrated with newly
discovered knowledge extracted from BN which was
trained on existing data set. In (Huhns 2007)
ontology is used for management of evidence in the
BN. In (Wang 2008) ontology is used for integration
of heterogeneous data sources and BN is used for
making probabilistic suggestions.
In medical domain, (Jeon 2007) uses ontology
for semi-automatic construction of BN for
diagnosing diseases. In (Zheng 2005) guideline
modelling tool that uses ontological workflow
management (GLIF) is integrated with probabilities
obtained by BN.
As we have demonstrated in this brief overview,
previous attempts of integration of these knowledge
representation formalisms are focused mainly on
calculation of probabilities of outcomes of some
events. Methodology proposed in this paper is
focusing mainly towards the IG of possible actions.
This is the crucial difference of the proposed
methodology with already existing approaches of
integration of ontologies and BNs.
3 USING BAYESIAN NETWORK
It is possible to construct BN (1) manually by
knowledge acquisition (in interaction with medical
experts), (2) automatically by machine learning
algorithms (from available medical data sets), where
it is possible to learn network structure and
conditional probabilities separately.
The machine learning approach is especially
useful in the medical domain where it is very hard to
explicitly state medical knowledge, and on the other
hand there already are plenty of available medical
data sets. With arrival of new patient data the
network can be updated and improved. If the
environment of the network is changed (e.g. the
system is applied in another country), new network
can be obtained by learning on new data set.
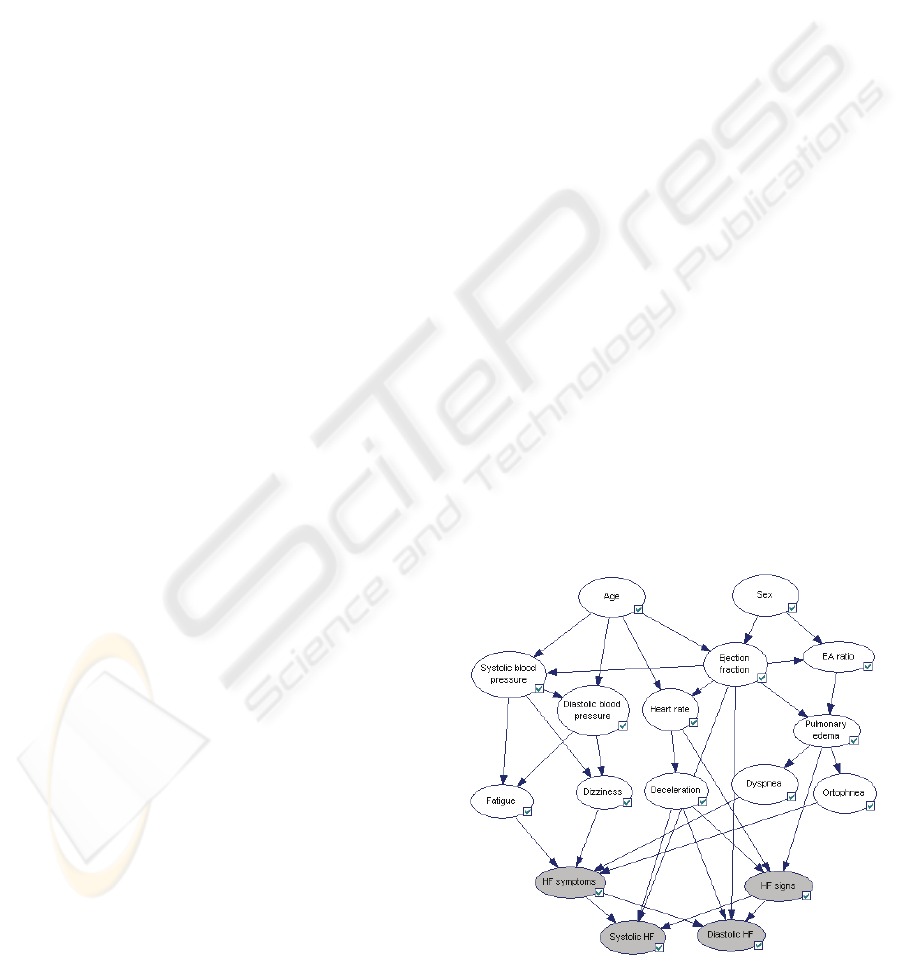
Figure 1 shows a provisional example of BN that
was built manually and that is used in the paper for
methodology demonstration purposes (from heart
Figure 1: Provisional BN for diagnosing heart failure
disease.
HEALTHINF 2010 - International Conference on Health Informatics
236
failure domain). Based on defined conditional
probabilities it is possible to calculate probabilities
(beliefs) of each outcome of each node in the
network (e.g. expecting normal ejection fraction in
70% cases).
In cases when physician is uncertain about the
diagnosis she should perform additional diagnostic
tests. In that case it would be very useful to know
which medical tests are the most appropriate in
currently observed patient situation. In other words,
it is useful to calculate IG of each observation node
for each target node. There are many measures
which could be appropriate in this situation; the
expected decrease of entropy is a measure which is
most commonly used (Jagt 2002).
To calculate the entropy of the target node we
use the probabilities of all outcomes of the target
node:
oo
o
ppXEXEntropy
2
log)()(
∑
−==
where X is a target node and o is the outcome of the
target node. The maximum value of entropy is 1
(when considering only two possible outcomes: Y
ES
and N
O) and it is reached when the information
about the target node is completely uncertain (when
P(Y
ES) = P(NO) = 0.5). As probability of target node
approaches towards ends (0 and 1) the entropy is
falls into zero. It is better to have the entropy values
as close to zero as possible since that indicates that
the answer to the target question is clearer.
A summary measure which takes all possible
outcomes of diagnostic test into account is called
expected entropy which is calculated as follows:
),|(),( dDXEpDXtropyExpectedEn
d
d
==
∑
where X is target node, D is observation node, d
is a single outcome of node D, p
d
is probability of
occurrence of d outcome, and E(X|D=d) is the
entropy of the target node X when outcome d has
happened. It can be shown that in any BN value of
expected entropy cannot be raised by any diagnostic
test, only lowered (Jagt 2002).
When the procedure described above is repeated
for all observation nodes, a diagram shown on
Figure 2 is obtained. The figure indicates that for
reaching the final decision whether patient has or
has not diastolic heart failure the most informative
diagnostic tests is measuring diastolic blood
pressure. After physician actually measures diastolic
blood pressure beliefs in the network are updated.
Accordingly, observation nodes are updated with
fresh IG values. Such reasoning procedure in BN is
referenced in the literature as “explaining away”.
Figure 2: IG of all diagnostic nodes for target concept
D
IASTOLICHF, for a patient that has not performed a single
diagnostic test yet.
4 STRUCTURED MEDICAL
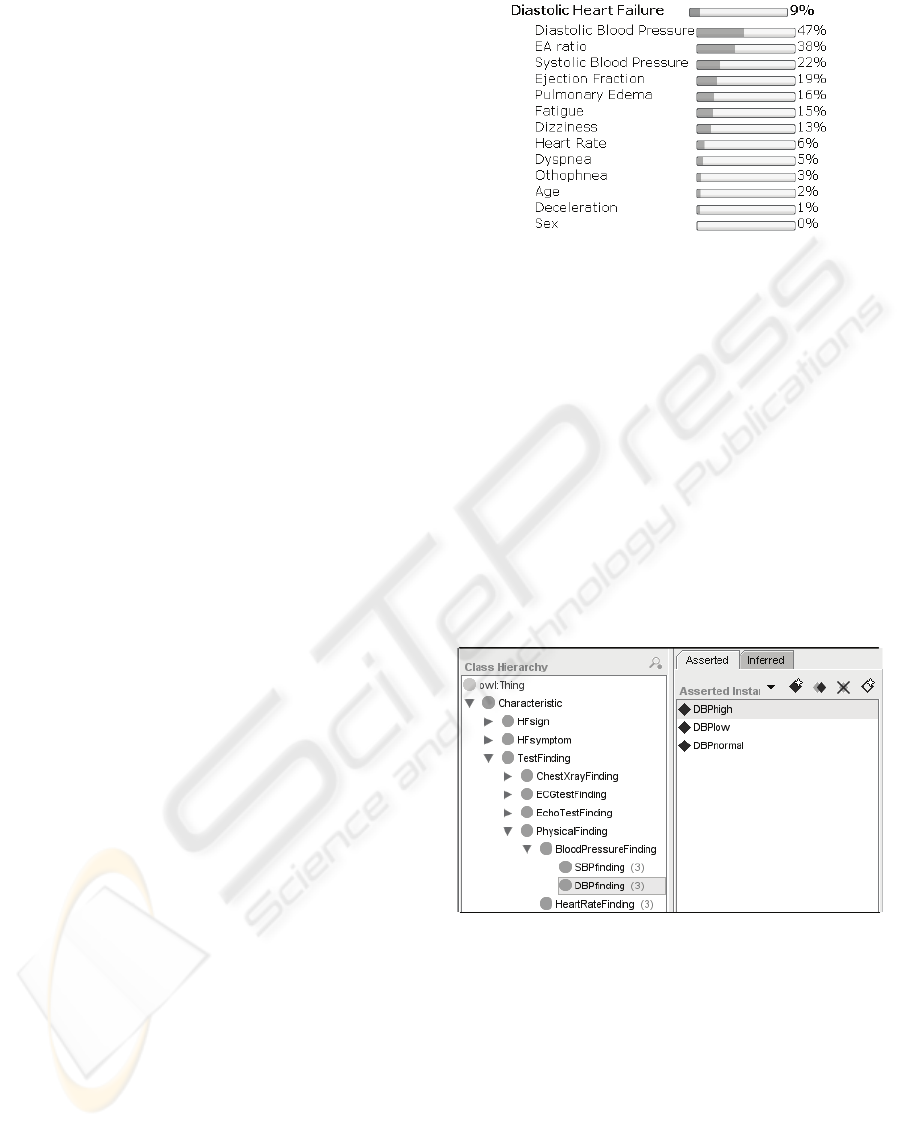
DIAGNOSTIC TESTS
In medical practice some diagnostic measurements
are never performed separately (e.g. systolic and
diastolic blood pressure). Ontology provides a
framework for organizing all possible diagnostic
tests into groups as they appear in medical practice.
In the ontology grouped diagnostic tests are
organized easily by arranging the ontology structure
as shown on Figure 3.
Figure 3: Diagnostic tests are defined within ontology.
The figure shows which elementary diagnostic
values are measured by performing blood pressure
measurement grouped diagnostic test. Besides
identifying blood pressure measurement as an
independent test, it is at the same time a constituent
part of a more thorough tests which is called
physical examination. In this manner it is possible to
organize diagnostic tests into groups and subgroups.
Additionally, some elementary observation can be a
part of two different tests; e.g. heart rate can be
measured both on physical examination and on
INFORMATION GAIN OF STRUCTURED MEDICAL DIAGNOSTIC TESTS - Integration of Bayesian Networks and
Ontologies
237
ECG. Grouped diagnostic tests are not necessarily
disjunctive.
4.1 Outcomes of Structured Tests
Difficulty with structured diagnostic tests is that the
number of possible outcomes grows extremely fast.
Namely, it is equal to the product of number of
outcomes for every elementary test in the group. For
example, if group blood pressure measurement has
only two elementary measurements (systolic and
diastolic) where each has three possible outcomes
(high, normal, low), there are nine possible
outcomes of such test. It is evident that the growth
rate of total number of outcomes in the group is of
combinatorial nature.
4.2 Information Gain of Structured
Medical Test
Formally speaking, to calculate the IG of a group for
every possible outcome g of the group G one must
calculate (1) the a priori probability of occurrence of
observed group outcome p
g
, and (2) entropy of the
target concept when observed outcome g happens E
g
= E(X | G = g). Then the expected entropy of the
target concept is equal to:
∑
=
g
gg
EpGXtropyExpectedEn ),(
where X is observed target node and G is observed
grouped diagnostic test.
To find exact value of probability p
g
it is possible
to construct a dummy node in the BN which would
have all nodes from the observed group as parent
nodes and conditional probability table defined as
truth table which evaluates to Y
ES only in the
column of the observed outcome g. When network
beliefs are updated the belief of outcome Y
ES in that
node will be equal to p
g
. By setting the evidence on
the same dummy node to the outcome YES one
could read out the a posteriori probability of the
observed target node and thus calculate value E
g
.
This procedure should be repeated (1) for every
possible outcome, (2) of every possible diagnostic
group test, and (3) for every possible target node.
With large number of target nodes, large number of
grouped diagnostic tests and large number of
possible outcomes the procedure becomes extremely
computationally demanding. This calls for other
potential solutions which would compute in more
acceptable time.
One possibility for solving this issue is to use the
sampling algorithms. Namely, it is possible to
generate arbitrarily large set of samples (artificial set
of patients) depending on the properties of the BN
and depending on patient evidence that is present
and to use it to calculate required probability values.
The same procedure applies with grouped tests:
samplesofnumber
goutcomewithsamplesofnumber
p
g
=
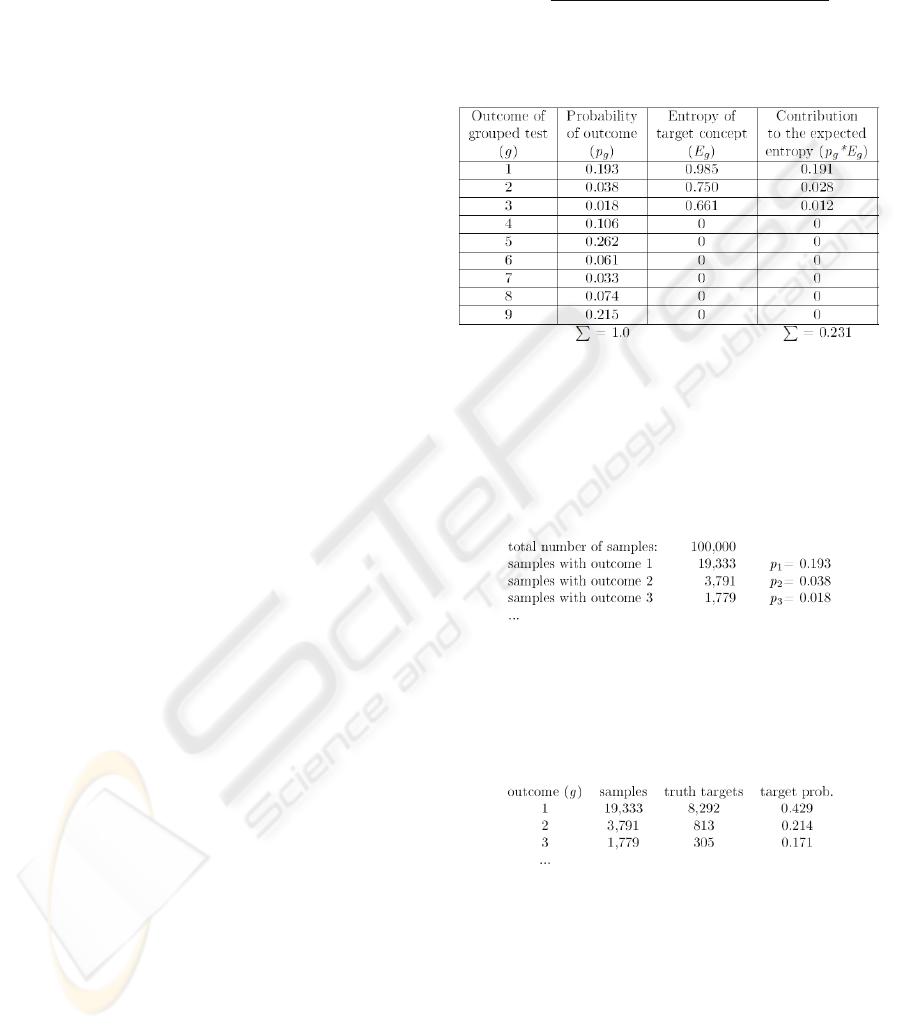
Table 1: Calculating the expected entropy of the target
concept with respect to the grouped diagnostic test.
Table 1 explicates the procedure for computing
the expected entropy (0.231) of the target node
(D
IASTOLIC HF) after performing grouped diagnostic
test (measuring blood pressure). By counting the
number of samples with observed outcome one
calculates probabilities of outcomes p
g
(second
column of Table 1):
This way the initial set of samples has been
divided (unevenly) into nine disjoint subsets. In each
subset it is possible to count samples for which the
target node was assigned with positive diagnosis.
This way a posteriori target probabilities are also
calculated from the same sample set:
Now it is also possible to calculate the a
posteriori entropy values and also the final IG value.
5 USAGE EXAMPLE
By starting the decision support services physician
finds out probabilities of diseases for the observed
patient considering all currently known patient data.
An example is shown on Figure 4. When the
analysis of IG for all defined grouped diagnostic
HEALTHINF 2010 - International Conference on Health Informatics
238
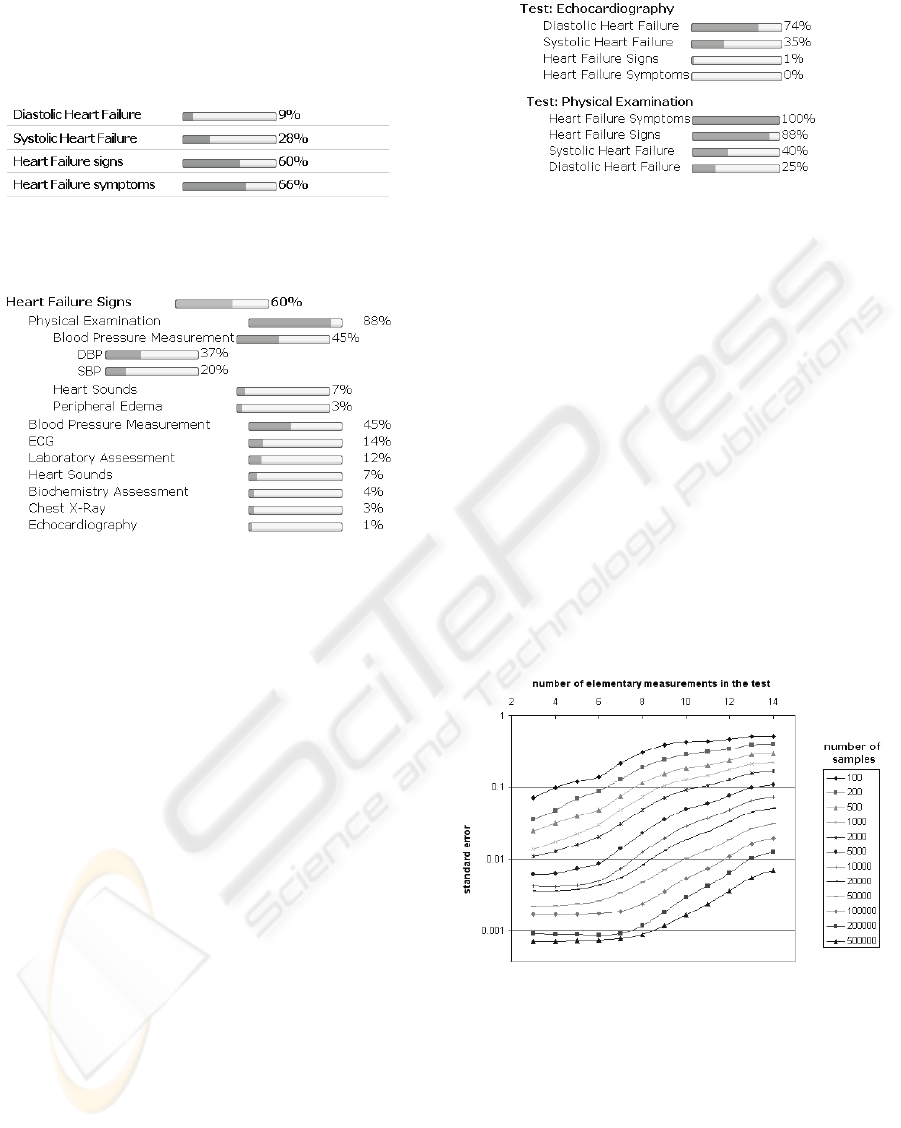
tests in the ontology is performed, the physician can
find out which test is the most informative one
considering observed target nodes. Figure 5 shows
the example results of such analysis.
Figure 4: Probabilities of target nodes for observed patient
considering all currently known patient data.
Figure 5: IG of ontologically organized diagnostic tests
considering the target concept “Heart failure signs”.
Medical interpretation of results shown on Figure
5 is following: considering all his currently known
data and considering all previously recorded cases of
the disease (cases which are inherently encoded
within BN) observed patient has 60% possibility of
having heart failure signs. If physician wants to be
more certain he can perform some additional
medical tests. Analysis of IG (again, considering all
known patient data and considering all previously
recorded cases) indicates that physical examination
is the most informative test that can be performed.
Physical examination contains more diagnostic
tests one of which is measuring blood pressure,
which can be further divided into measuring
diastolic and systolic blood pressure (structure
defined within the ontology). All such tests (both
grouped and elementary) have their own IG value.
A different view of the results is also possible:
one can compare the summary impacts of grouped
diagnostic tests on defined target concept. Figure 6
demonstrates the comparison of two diagnostic tests:
echocardiography and physical examination. This
way physician can compare the overall values of IG
of all medical tests which helps him to make a
decision which medical test (tests) should be
performed next.
Figure 6: Comprehensive view of IG of available medical
diagnostic tests.
6 PERFORMANCE
Behaviour of the system in a great deal depends
upon some inherent characteristic both of the
ontology and the BN. Within this paper
measurements are conducted using a single specific
BN and a single specific ontology; hence, the
analysis is merely a preview of some provisional
setting. However, we assume that behaviour of a
single problem instance at least to some extent
indicates its general behaviour.
The main concern in the performance of the
described system is with (1) time required for
reasoning and (2) error made in reasoning.
Furthermore, it is evident that there is a trade-off
between those two parameters.
Figure 7: Standard error made in reasoning depends upon
number of elementary measurements in the test and the
number of samples.
Figure 7 shows the dependency of error made in
the reasoning process upon the number of
elementary measurements in the test and the number
of samples used. For example, if one is calculating
the IG of some grouped test which contains 10
elementary measurements using 50,000 samples
INFORMATION GAIN OF STRUCTURED MEDICAL DIAGNOSTIC TESTS - Integration of Bayesian Networks and
Ontologies
239
standard error made in calculations will be
somewhere near 1%.
Figure 8: The appropriate number of samples depends on
used number of elementary measurements in the tests and
on chosen error rate.
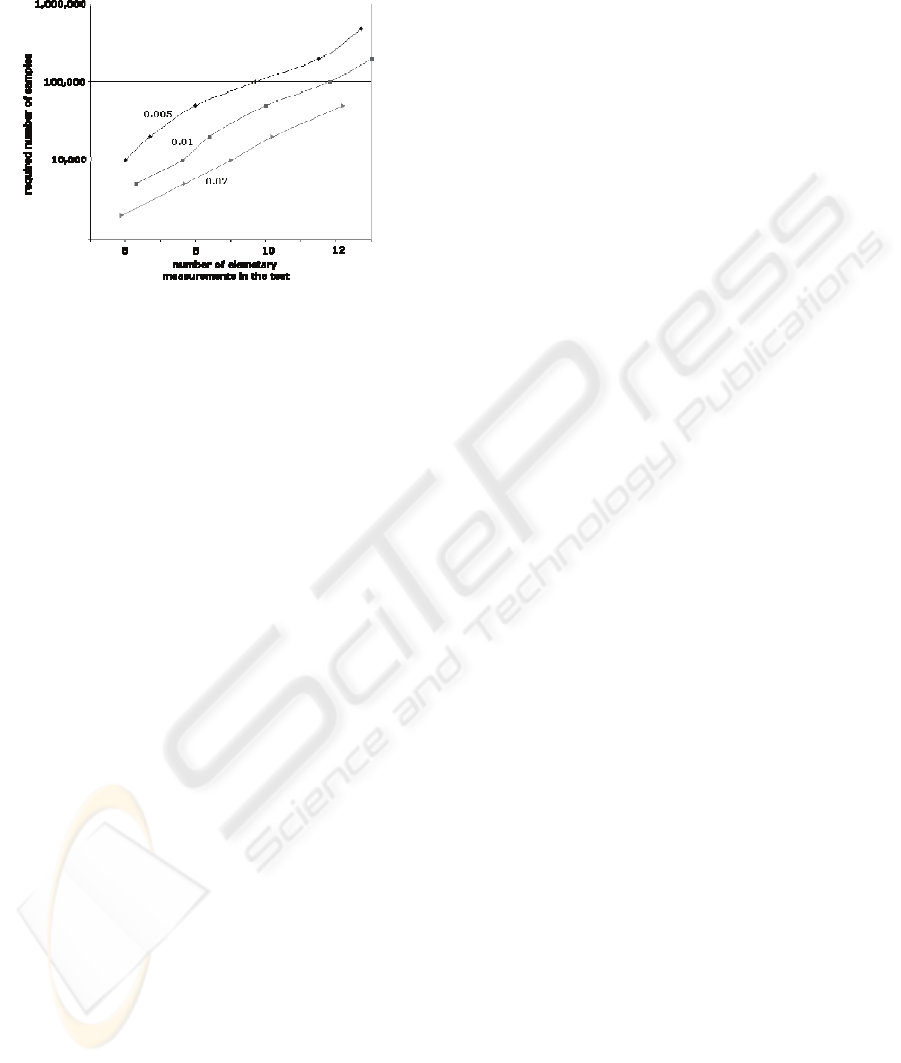
Figure 8 is indicating a minimum number of
samples one should use depending on sizes of
defined groups in the ontology and on chosen error
rate. E.g., if one is satisfied with error rate of 1% and
has up to 12 elementary measurements in a group
she should use at least 100,000 samples in reasoning
phase. Chart depicts such relation for error rates of
2%, 1% and 0.5%.
7 CONCLUSIONS
In this paper we have demonstrated the approach for
integration of knowledge from BNs and ontologies
in order to calculate the IG of structured medical
test. We strongly believe that the approach is sound
and can be very useful in practical medical decision
support systems.
The main obstacle in the described methodology
appeared to be the combinatorial nature of the
number of outcomes in grouped diagnostic tests.
However, practical experiments indicate that this
obstacle in some cases can be to some extent
avoided by usage of sampling algorithms in the
reasoning phase. The measurements have
demonstrated the dependency of error rate and
required number of samples. On that basis, and
considering some specific system properties such as
number of nodes in the network, sizes of grouped
diagnostic tests, acceptable time of reasoning,
acceptable error rate, and properties of machine that
performs reasoning, one can conclude which number
of samples should she use in the reasoning phase.
Structure of BNs inherently assumes conditional
independency – an assumption that in general case
does not stand for medical diagnostic tests. In spite
of that, vast majority of decision support systems
that make use of BNs ignore this issue. However,
one should be fully aware of this drawback when
using proposed methodology in practice.
Suggested methodology of integration of BNs
and ontologies still calls for more thorough testing
of its overall performance and has yet to
demonstrate its practical utility in real medical
environments. Furthermore, suitability of the
approach in some other domains remains to be
shown. All above mentioned problems seem to be
rather interesting topics for the future work.
REFERENCES
Pan, R., Ding, Z., Yu, Y., Peng, Y. (2005). A Bayesian
Network Approach to Ontology Mapping. Fourth
international Semantic Web Conf. 2005, 563-577.
Devitt, A., Danev, B., Matusikova, K. (2006).
Constructing Bayesian Networks Automatically using
Ontologies. Ontology Meet Industry workshop 2006.
Town, C. (2004). Ontology-driven Bayesian Networks for
Dynamic Scene Understanding. IEEE Computer
Society Conference on Computer Vision and Pattern
Recognition Workshops 2004.
McGarry, K., Garfield, S., Wermter, S. (2007). Auto-
Extraction, Representation and Integration of a
diabetes Ontology using Bayesian Network.
Computer-Based Med. Sys. Symposium ’07, 612-617
Huhns, M.N., Valtorta, M.G. (2007). Ontological Support
for Bayesian Evidence Management. Ontology for the
intelligence community 2007, 47-51.
Wang, W., Zeng, G., Zhang, D., Huang, Y. et al. (2008).
An intelligent Ontology and Bayesian Network based
Semantic Mashup for Tourism. IEEE Congress on
Services, part I, 128-135.
Jeon, B.J., Ko, I.Y. (2007). Ontology-Based Semi-
Automatic Construction of Bayesian Network Models
for Diagnosing Diseases in E-health Applications.
Frontiers in the Convergence of Bioscience and
Information Technologies, fbit, pp.595-602
Zheng, H.T., Kang, B.Y., Kim, H.G. (2005). An
Ontology-based Bayesian Network Approach for
Representing Uncertainty in Clinical Practice
Guidelines, Uncertainty Reasoning for the Semantic
Web workshop 2005.
Jagt, R.M. (2002). Support for multiple cause diagnosis
with Bayesian Networks. Thesis for degree of Master
of Science in Applied Mathematics. Information
Sciences department, University of Pittsburgh, USA.
HEALTHINF 2010 - International Conference on Health Informatics
240
