
TOWARDS THE EVOLUTION OF LEGACY APPLICATIONS TO
MULTICORE SYSTEMS
Experiences Parallelizing R
Gonzalo Vera and Remo Suppi
Computer Architecture and Operating Systems Department (CAOS), Universitat Aut`onoma de Barcelona
Campus UAB, Edifici Q, 08193 Bellaterra (Barcelona), Spain
Keywords:
Legacy applications, R language, Parallel computing, Multicore systems.
Abstract:
Current innovations in processor performance, focused to keep the growth rate of the last years, are mainly
based on providing several processing units within the same chip. With new underlying multicore processors,
traditional sequential applications have to be adapted with parallel programming techniques to take advantage
of the new processing capabilities. There exists a great variety of libraries, middlewares, and frameworks
to assist the parallelization of such applications. However, in many cases, specially with classical scientific
applications, due to several limitations ranging from technical incompatibilities to simply lack of knowledge,
this evolution cannot always be achieved. We here present our experiences providing an alternative for two
situations where former contributions could not provide a satisfactory solution to our needs: adapting a mature
non-thread-safe C coded application, the R language interpreter, and providing support for the automatic
parallelization of R scripts in multicore systems.
1 INTRODUCTION
During the last years we are witnessing a new age
characterized by massive adoption of multiprocessor
computers, even at desktop level with the populariza-
tion of multicore computers. The change on the gen-
eralized way microprocessor manufacturers are in-
creasing the raw performance of their products started
a debate still open about the serious implications of
these new micro-architectures over applications per-
formance (Sutter and Larus, 2005). There is a large
set of classical scientific applications that have been
extensively used since the last decades without taken
into consideration the underlying computer architec-
ture. An example of this type of applications are
bioinformatics software tools using statistical or ar-
tificial intelligence methods to analyze the results of
experimental data based on the scripting language R
(Ihaka and Gentleman, 1996). The R language inter-
preter, like many other legacy applications is a single-
thread program that is not prepared out of the box
to take advantage of nowadays multicore computers.
In order to do so parallel programming techniques
are required. As a consequence, legacy applications
like the R language interpreter, running on top of cur-
rent multicore processors, claim for parallel comput-
ing support.
Transforminga sequential program to make it able
to run concurrently several sections of its code it is a
difficult task that can be achieved by several meth-
ods, depending on our resources, requirements and
limitations (Bridges et al., 2008). Most of them im-
ply either using a shared memory model, suitable
for multiprocessor machines, or a message passing
model, which can be used between networked ma-
chines. Shared memory based solutions, within a sin-
gle process, make use of multithreading techniques
to run several instances of a single process together
with shared variables and inter-process communica-
tions (IPC) mechanisms like mutex sections to syn-
chronize its parallel execution. A useful tool to au-
tomate the construction of such programs is OpenMP
(Dagum and Menon, 1998). Solutions based on the
message passing paradigm also use IPC mechanisms
but in this case to communicate different processes,
usually but not necessarily, through the network. A
well-known library that provides a full set of building
blocks to ease the construction of such parallel pro-
250
Vera G. and Suppi R. (2010).
TOWARDS THE EVOLUTION OF LEGACY APPLICATIONS TO MULTICORE SYSTEMS - Experiences Parallelizing R.
In Proceedings of the First International Conference on Bioinformatics, pages 250-256
DOI: 10.5220/0002746302500256
Copyright
c
SciTePress

grams is MPI (MPI Forum, 1993). There are more
solutions that have proved their value with great suc-
cess, but even in the case these useful tools are com-
patible with our programming language and running
environment, more inconvenients still can appear.
An obstacle that dramatically increases the com-
plexity to adapt sequential programs appears when
global variables are extensively used in legacy appli-
cations. In these situations, if using multiple threads
running at different instructions of the same process,
it is quite complicated to ensure the correct access
to these variables and avoid race conditions. When
correct access order is not ensured, applications are
said to be non-thread-safe. This is quite common
in large legacy applications that have been growing
for years while increasing its funcionality. An ex-
ample of this, with more than 10 years of evolu-
tion, is the R language interpreter. A different prac-
tical obstacle appears when observing the mainte-
nance lifecycle of legacy applications. After years
of proved utility it is logical to expect a long life
span. Introducing an external dependency on a piece
of software that later on may get discontinued can
cause serious problems in the future. Another legal
obstacle is found between incompatible software li-
censes. For example, many open-source tools are
published using the GNU General Public License
GPL (GNU Software Foundation, Inc., 2007). Al-
though partially solved with the Lesser GPL license,
the former prevents the usage of these GPL licensed
tools together with propietarysoftware licenses which
were commonly adopted by earlier legacy applica-
tions. Finally, a pragmatic problem comes with the re-
quired skills to perform such transformations or adap-
tations. It is common for scientists to program their
own applications. Although when implementing their
algorithms, they produce high quality applications,
without specific background on software engineering
and parallel computing, this process of transforma-
tion, due to the lack of knowledge and experience, is
very cumbersome and error-prone.
In this paper we expose our research experiences
creating a solution to support multicore systems in
the R language. The outcome is an add-on R pack-
age called
R/parallel
(Vera et al., 2008). Its con-
ception initiates after the need of providing parallel
support for the R language interpreter, a non-thread-
safe C coded legacyapplication. The next sections de-
scribe the reasons and motivations that have directed
our design decisions and implementation details in or-
der to allow other developers, with equivalent needs
and conditions, to adopt a similar solution. Besides
of providing a technical description of an explicit par-
allelism method to enable the usage of multicore pro-
cessors and assist other legacy application maintain-
ers, we also describe with more detail our extension of
the R language interpreter, including the experimental
results that have allowed us to validate our implemen-
tation. This second contribution also provides assis-
tance to enable parallel computing, but in this case
providing a simple parallelization method that any R
user, using an implicit parallelism method, and with-
out aditional programming skills, can use to run his or
her R scripts.
2 DESIGN CONSIDERATIONS
The design of a solution to provide parallel computing
capabilities in single thread legacy applications like
the R language interpreter is constrained by the prob-
lems exposed previously in the introduction section.
Language interpreters are a good example of pro-
grams that have evolve considerably over time. Their
implementations can be grouped into two different
approaches observing how they handle the global
variablesshared between differentconcurrentthreads:
share all or share nothing. The share all approach
has the advantage that any variable is directly ac-
cessible at any time. However, since the access has
to be controlled continuously with global locks, its
performance falls down with an increasing number
of parallel threads. The second approach, in con-
trast, shares nothing unless explicitly defined. This
imposes more work for the programmer but results
in better scalability. This approach has been used
by many language interpreters like python, erlang
or perl. In fact, the perl implementation of threads
switched from the share all to the share nothing ap-
proach in version 5.8.0 (The Perl Foundation, 2002)
to overcome the poor performance of its earlier im-
plementations. With this second implementation the
scalability is dramatically increased although the se-
quential access to shared variables is still a bottleneck.
The interpreter implementations, besides of
choosing a share nothing approach, can be classified
into two additional groups. One group implements
dedicated user level threads to manage the shared re-
sources, also known as green threads, while the other
delegates its control to native system calls at kernel
level, known as native threads. The first option has
the advantage, on single processor computers, that
since the controlling thread has specific knowledge
about their own family threads, its expected perfor-
mance should be greater than if managed by general
purpose kernels, which have no knowledge about the
future requirements of the threads being scheduled.
However, the common disadvantage, since all user-
TOWARDS THE EVOLUTION OF LEGACY APPLICATIONS TO MULTICORE SYSTEMS - Experiences Parallelizing
R
251
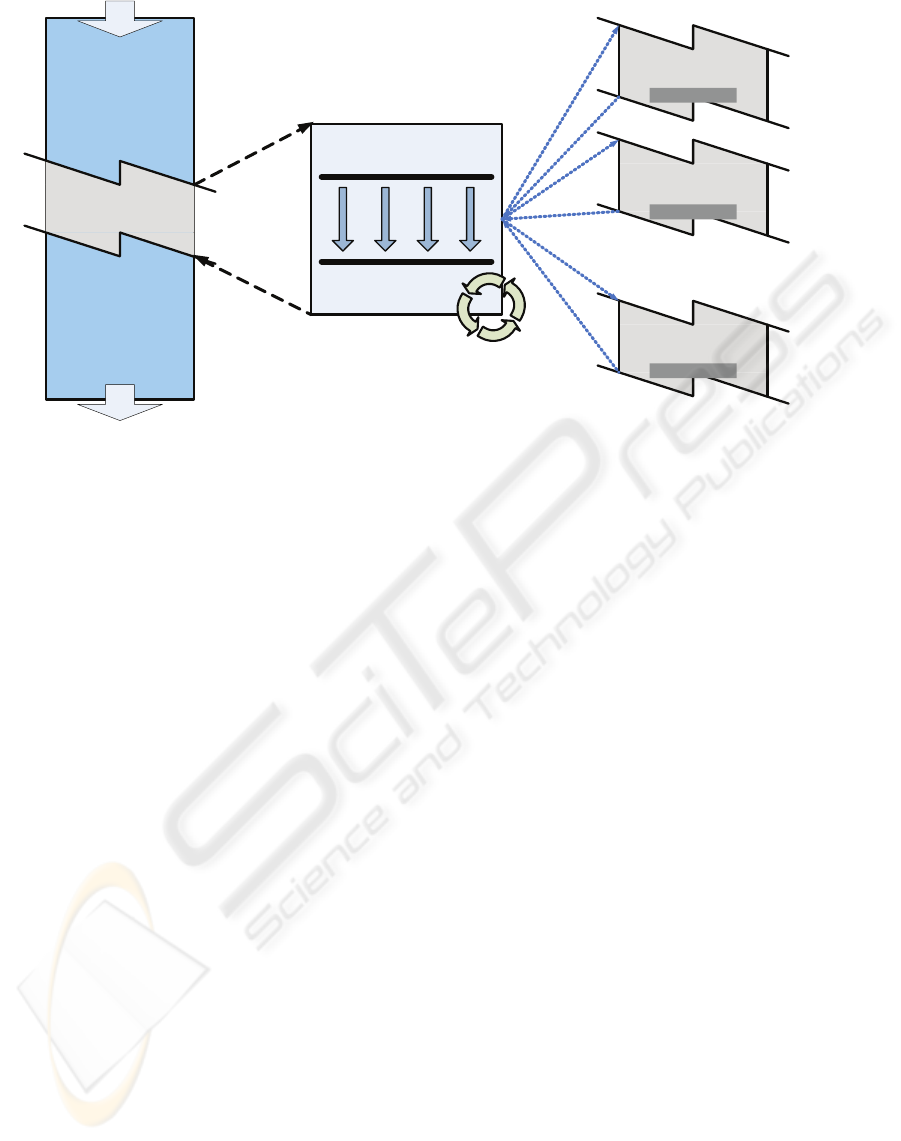
parallel
section
sequential
section
sequential
section
parallel
section
parallel
section
parallel
section
.
.
.
INSTANCE #1
INSTANCE #2
INSTANCE #N
Independent processes
(running different instances of the parallel section )
Added
Multithreading ModuleLegacy Application
SET UP, DISTRIBUTION
and COORDINATION
COLLECTION &
REDUCTION
Re-entrant code
Figure 1: General design strategy for parallelizing a non-thread-safe legacy application.
level threads belong to the same process and they
share the processor quantum of scheduled time (i.e.
cooperative timeslicing scheduling) is that only one
thread is scheduled at a time to a processing unit.
With a high number of running threads, the control-
ling thread turns into the busiest one, blocking the
others to get access not only to the shared variables
but also to their share of processor time. This scala-
bility problem appeared for example in early versions
of the java virtual machine or in the ruby interpreter
language. Using native threads have been adopted by
other programs like the python interpreter or later ver-
sions of the java virtual machine to solve this limita-
tion. Taking into account the evolution and experi-
ence of those general purpose interpreters seems logi-
cal that when performance on multicore systems mat-
ters, and restricted to the situations depicted in the in-
troduction, using a share nothing approach based on
internal operating system mechanisms is the recom-
mended option.
Being the R language interpreter a considerably
large non-thread-safe application it is not advised to
initiate a restructuration that will require an exten-
sive revision of all the global variables used all over
its source code and a later validation of the changes
introduced to ensure its initial quality levels. More-
over, the R language interpreter, like many modern
languages, is evolving continuously, and every year a
few updates are released. That will require a contin-
uous tracking of the changes introduced in new ver-
sions that clearly discourages any direct modification.
At the other hand, choosing a third party tool, if avail-
able and compatible, technically and legally, for our
application, has to be carefully done if we expect this
introduced dependency to exist safely for the coming
years. Therefore, as long as multithreading within the
same working process is not directly a feasible op-
tion, a classical alternative, multiprocessing, seems a
right choice. The requirement for that option is to
find a way to create multiple processes with selected
code and manage its execution. As we exposed ear-
lier, libraries like MPI provide helpful functions than
can assist with the task of spawning and communicat-
ing processes but they also have two major inconve-
nients for our specific needs: first, they require the in-
stallation and configuration of additional system soft-
ware, what turns to be too difficult and scary for non-
technical users, and second, the available wrappers
existing for the case of R does not provide an standard
and stable programming interface over MPI versions
(e.g. Rmpi (Yu, 2009) does not have a seamless inte-
gration of different MPI implementations like LAM
(Burns et al., 1994) and OpenMPI (Gabriel et al.,
2004)).
Finally, taking all the arguments into considera-
tion, the design chosen is depicted in figure 1. The
basic idea is to identify, within the legacy applica-
tion the sections of code that can independently run
in parallel. These sections can be replicated in several
independent processes so we are sure we will avoid
race conditions when accessing its local copy of the
global variables. In order to prepare these processes
with different input data, coordinate its execution and
collect back the partial results we need a central piece
BIOINFORMATICS 2010 - International Conference on Bioinformatics
252
of software. This additional module, as long as it is
completely new and shares nothing with the original
application can be implemented using multithreading.
These threads will be used to manage independently
the creation and communication of the processes with
the module. The result is a master-worker architec-
ture, suitable for embarrassingly parallel problems,
where the central module acts as a master coordinator
and a set of working processes, running concurrently
over different processor cores, perform the calcula-
tions that previously were done sequentially within
the legacy application.
3 IMPLEMENTATION
Following the design described in the previous section
we have implemented an R add-on package to extend
the functionalities of the R language interpreter and
provide support for parallel computing in multicore
systems. The implementation mixes standard R core
functions to interface with the R interpreter (and the
R user), together with C++ objects to interface with
the operating system and manage the parallelization.
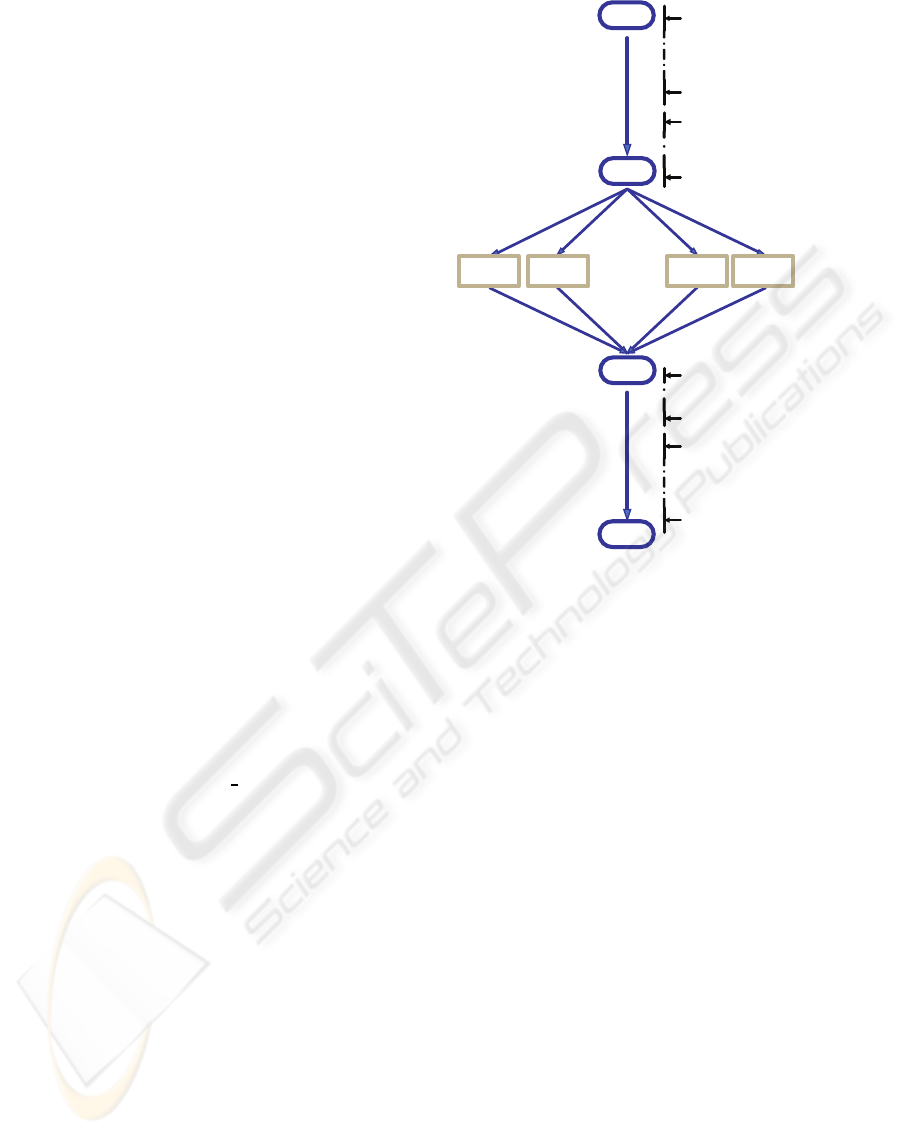
The general steps undertaken by the master module
are summarized in figure 2. Once a parallel section is
reached within the legacy application, the execution
is diverted to the added controlling module where the
first step performed is to retrieve the current value of
all the accessible variables of the ongoing execution.
With this information, and knowing the parallel sec-
tion of code to be run, the independent processes (jobs
in advance) are set up and spawned using bootstrap
files and system calls (i.e. fork like functions). Using
standard system calls, although less straightforward
than using already done wrapper libraries ensures the
autonomy, and therefore the long term maintainabil-
ity of the application. At this point, each worker will
perform its assigned job and once finished it will be
returned to the master. The communication is carried
out and coordinated using standard IPC system calls
and objects (i.e. mutex variables and pipes).
Once all the partial results are recovered, and
knowing the jobs assigned to each worker, the mas-
ter module is able to compute its aggregated results
(i.e. reduces the partial values) to obtain the single
final values. At this point, the modified variables are
updated within the legacy application, and the execu-
tion continues from the next sequential section with-
out further changes.
With this strategy, we can effectively run concur-
rently any section of R scripts by rising several in-
stances of R conveniently prepared to communicate
with the central module. However, how R end-users
0. Initial sequential execution
1. Save the state
2. Create the jobs
3. Submit the jobs
. . .
external
execution
4. Retrieve the results
5. Compute aggregated result
6. Final sequential execution
7. Return to invoking call
worker
process
worker
process
worker
process
worker
process
Figure 2: Steps sequence performed at the master module.
can take advantage of this mechanism still have to be
defined. The solution implemented in our proposal is
illustrated in figure 3.
Since loops without data dependencies (i.e. paral-
lel loops) are the most common case within bioinfor-
matics applications, where large vectors of samples
are analized with the same analytical method, one af-
ter the other, this is the first situation where automatic
parallelization can bring great benefits, and therefore
the first that our implementation currently supports.
Once an R user has coded his or her function with a
for
loop, to run its iterations concurrently, he only
needs to enclose the
for
loop within the
else
body
of an additional
if-else
conditional structure. In the
case of being our R add-on package loaded, the par-
allelization will take place. Otherwise, the execution
will run as usual without any change. This method,
instead of using directly a new external function call,
and as far as R does not provide macros (used in tools
like OpenMP), has been chosen to let R users to keep
sharing their scripts, regardless of using or not our R
add-on package. If the
if
condition is true, then the
function
runParallel()
can safely be called. At this
point, the original thread of execution is diverted to
the master module. There, all the accesible variables,
including the
for
loop code of the invoking function,
together with the
runParallel()
arguments, are re-
trieved using the scoping functionalities of R. This
TOWARDS THE EVOLUTION OF LEGACY APPLICATIONS TO MULTICORE SYSTEMS - Experiences Parallelizing
R
253
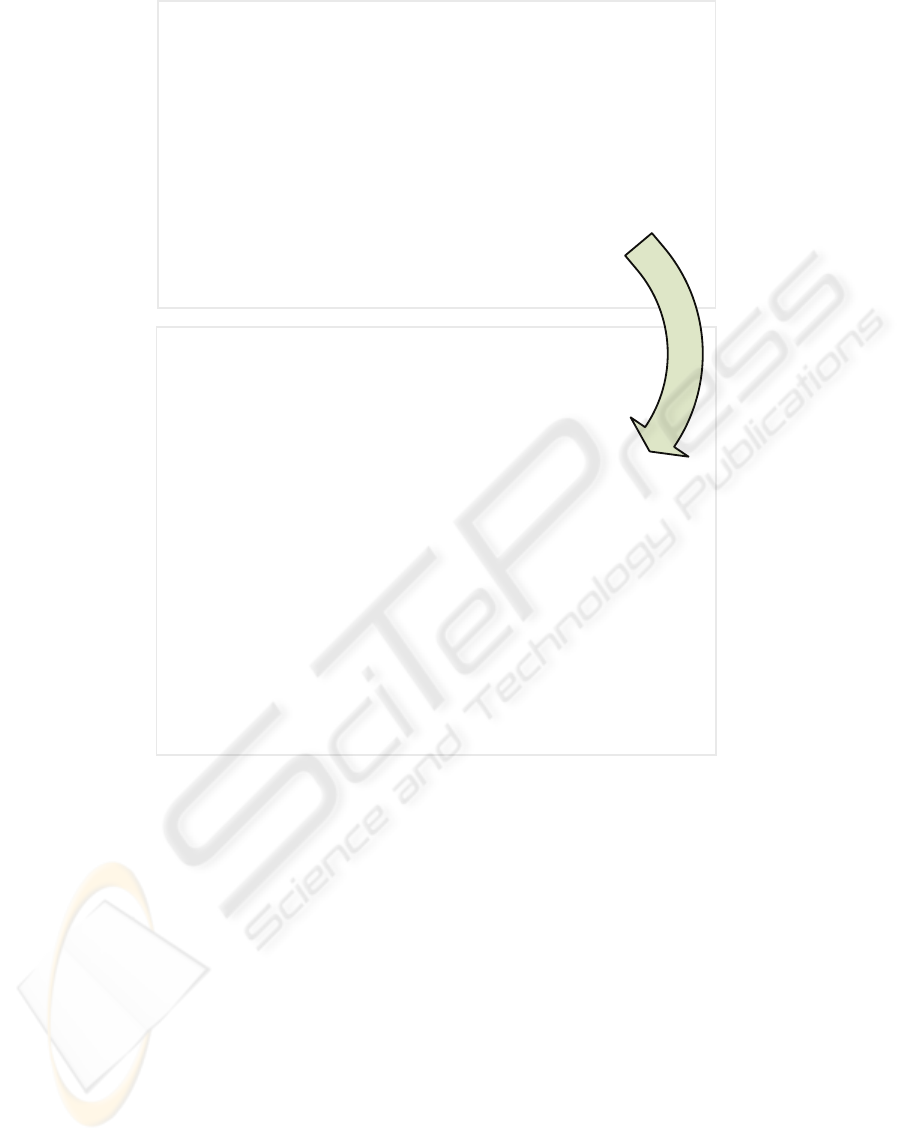
yourFunctionName <- function( argument1, argument2=NULL )
{
# 1. Initializing Variables
anyVar <- 0
reduceVar <- NULL
if( "rparallel" %in% names( getLoadedDLLs()) )
{
runParallel( resultVar="reduceVar", resultOp="reduceOp" )
}
else
{
# 2. Start of loop
for(index in 1:nrow(argument1))
{
#Make some calculations
internalVar1 <- someCalculations( argument2 )
tempResult <- someOperations(argument1[index], anyVar )
reduceVar <- reduceOp( tempResult, reduceVar )
}
}
# 3. Finalizing the function
return( reduceVar )
}
yourFunctionName <- function( argument1, argument2=NULL )
{
# 1. Initializing Variables
anyVar <- 0
reduceVar <- NULL
# 2. Start of loop
for(index in 1:nrow(argument1))
{
#Make some calculations
internalVar1 <- someCalculations( argument2 )
tempResult <- someOperations( argument1[index], anyVar )
reduceVar <- reduceOp( tempResult, reduceVar )
}
# 3. Finalizing the function
return( reduceVar )
}
Indicate Parallel Region
Figure 3: Generic example to parallelize an R loop.
step can also be programmatically accomplished in
any legacy application in case we have access to its
source code. After retrieving this information, the
master module begins the steps described previously.
The mandatory arguments of
runParallel()
are the
reduction variables which values have to be preserved
between iterations, and its corresponding reduction
operations used to aggregate the partial results. Fi-
nally, once the calculation has concluded, from the
same
runParallel()
function, the R environment of
the invoking function is updated with the new values
of the reduced variables. From this point, the execu-
tion of the R interpreter, or any other legacy appli-
cation where the same steps have been implemented,
will continue running the remaining sequential code.
4 EXPERIMENTAL RESULTS
In order to assess the capabilities of our proposal, to
take advantage of the available processing power of a
multicore computer when running a parallel comput-
ing extension of the R language interpreter, we show
in this section the results obtained after performing a
set of experiments selected for this purpouse.
The tests performed have been done using the
function
qtlMap.xProbeSet()
from the R add-on
package
affyGG
(Alberts et al., 2008).
affyGG
has
been developed to perform bioinformatics QTL anal-
ysis of samples obtained using Affymetrix microar-
rays. The input data has been simulated using real
data obtained from samples of 30 recombinant inbred
mice (Bystrykh et al., 2005) to obtain a total execution
time of the R function of +10 hours without using any
BIOINFORMATICS 2010 - International Conference on Bioinformatics
254
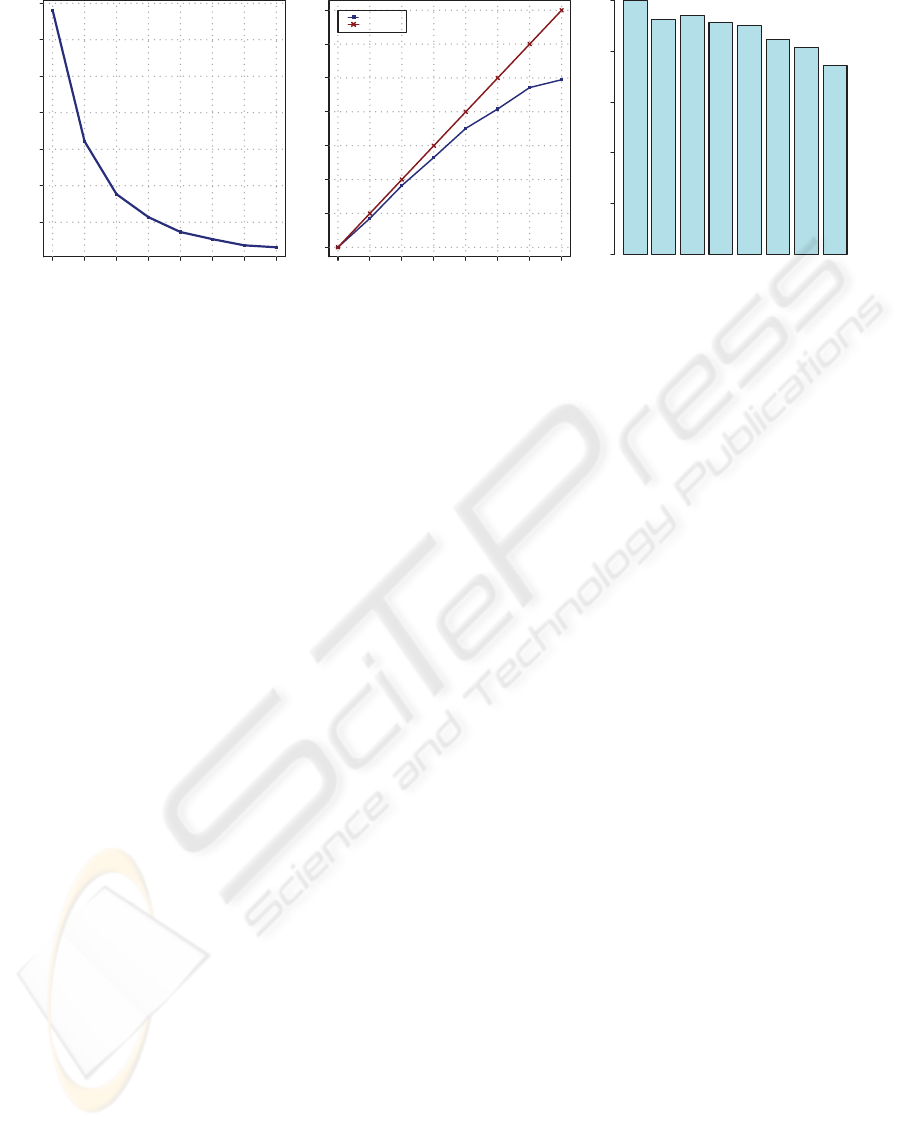
10000 15000 20000 25000 30000 35000 40000
Number of cores
Execution Time (seconds)
Number of cores
Speedup
Real
Linear
0.0 0.2 0.4 0.6 0.8 1.0
100%
92%
94%
91%
90%
84%
81%
74%
Number of cores
1 2 3 4
5 6 7
8
1 2 3 4
5 6 7
8
1 2 3 4
5 6 7
1 2 3 4
5 6 7
8
8
Eciency
Figure 4: Experimental Results.
parallel solution. By this way, adding progressively
more cores to the computation (the number of workers
can be set optionally), when running with our solution
we can observe how the scalability and efficiency of
this solution evolve as we add more cores.
The test environment consist of one server
equiped with 2 quad-core processors (i.e. 8 cores
available) and 16 GB of main memory running the
operating system Red Hat Enterprise Linux Server re-
lease 5 and the R language interpreter, version 2.8.1.
Figure 4 shows the results obtained of total execution
time, speedup and efficiency.
As it can be observed, the processing time is re-
duced proportionally as we add more cores to the
computation. Looking at the speedup, although ini-
tially close to the linear speedup, it is clear that, be-
cause of the overhead introduced with the manage-
ment and control the parallel execution, increasing the
number of processing units, the performance growth
rate is affected negatively. The less efficient case is
observed when using 8 cores. Besides of the system
processes, we have also to take into account the mas-
ter process. When reaching the maximum number of
available cores the machine is overloaded because of
the competence between all the processes trying to
get their corresponding slice of processor time. As a
consequence, the overall performance is affected and
the results, although still reducing the total execution
time, show a worse efficiency using 8 cores than using
other smaller configurations.
Nevertheless, the results demonstrate that even
with few available cores, our proposal, by enabling
the available computational power of nowadays mul-
ticore processors, and with so little effort by the R
user, is able to run parallel loops in R scripts substan-
tially faster than previously without our extension.
5 CONCLUSIONS AND FUTURE
WORK
In this manuscript we have described our experiences
parallelizing R from two points of view. One describ-
ing our experiences when parallelizing a non-thread-
safe legacy application by extending the R language
interpreter, and another, describing the characteristics
and benefits of using our R add-on package from an
end-user point of view. The final outcome is an R add-
on package called
R/parallel
which can be loaded
dinamically into the R language interpreter and allows
the parallel execution of
for
loops without data de-
pendencies using the strategy explained previously.
The design principles have been proved correct re-
garding the supporting technologies chosen. The R
package has remained completely independent and
functional across several version updates of R since
R/parallel
was released for the first time. Regard-
ing its perfomance benefits, the experimental results
show that our proposal enhaces the efficiency with
which R natively runs on top of multicore systems.
However, new functionalities can be implemented
and best performance be achieved. Current imple-
mentation is limited to
for
loops. Although this is
enough for most R users, there are other situations
where parallelism can be exploited. For example from
series of consecutive independent heavy-load func-
tion calls where each call can be performed by dif-
ferent workers (i.e. task parallelism). Increased per-
formance should also be achieved by controlling the
cores to which each worker is assigned. By control-
ling the process afinity it is possible to make a better
use of the processor cache memory and hence reduce
the execution times. Moreover, further performance
can be achieved by means of distributed computing.
This is an interesting research aspect for future work
TOWARDS THE EVOLUTION OF LEGACY APPLICATIONS TO MULTICORE SYSTEMS - Experiences Parallelizing
R
255

although extending the computing environment to re-
mote computers also increases the number of prob-
lems to deal with like for example load distribution
and fault tolerance.
Nevertheless, we expect our experiences will help
other legacy applications in the same situation de-
scribed for the R language. In such situation it is
faster to evolve coding and testing a new small mod-
ule than reviewing, restructuring and testing again the
whole body of very large applications. The same con-
cept applies for R end-users. Now, with our contribu-
tion, they are able to, in less time than before, quickly
and easily parallelize the execution of their
for
loops.
ACKNOWLEDGEMENTS
This research has been supported by the MEC-
MICINN Spain under contract TIN2007-64974.
REFERENCES
Alberts, R., Vera, G., and Jansen, R. C. (2008). affyGG:
computational protocols for genetical genomics with
Affymetrix arrays. Bioinformatics, 24(3):433–434.
Bridges, M. J., Vachharajani, N., Zhang, Y., Jablin, T., and
August, D. I. (2008). Revisiting the sequential pro-
gramming model for the multicore era. IEEE Micro,
28(1):12–20.
Burns, G., Daoud, R., and Vaigl, J. (1994). LAM: An open
cluster environment for MPI. In Proceedings of Su-
percomputing Symposium, pages 379–386.
Bystrykh, L., Weersing, E., Dontje, B., Sutton, S., Pletcher,
M. T., Wiltshire, T., Su, A. I., Vellenga, E., Wang,
J., Manly, K. F., Lu, L., Chesler, E. J., Alberts, R.,
Jansen, R. C., Williams, R. W., Cooke, M. P., and
de Haan, G. (2005). Uncovering regulatory pathways
that affect hematopoietic stem cell function using ’ge-
netical genomics’. Nature Genetics, 37(3):225–232.
Dagum, L. and Menon, R. (1998). OpenMP: An industry-
standard API for shared-memory programming. IEEE
Computing in Science and Engineering, 5(1):46–55.
Gabriel, E., Fagg, G. E., Bosilca, G., Angskun, T., Don-
garra, J. J., Squyres, J. M., Sahay, V., Kambadur, P.,
Barrett, B., Lumsdaine, A., Castain, R. H., Daniel,
D. J., Graham, R. L., and Woodall, T. S. (2004). Open
MPI: Goals, concept, and design of a next generation
MPI implementation. In In Proceedings, 11th Euro-
pean PVM/MPI Users Group Meeting, pages 97–104.
Ihaka, R. and Gentleman, R. (1996). R: A language for data
analysis and graphics. Journal of Computational and
Graphical Statistics, 5(3):299–314.
GNU Software Foundation, Inc. (2007). GNU general pub-
lic licence. http://www.gnu.org/licenses/gpl.html.
MPI Forum (1993). MPI: A Message Passing Interface. In
Proc. of Supercomputing 93, pages 878–883.
The Perl Foundation (2002). Perl 5.8.0 release announce-
ment. http://dev.perl.org/perl5/news/2002/07/18/
580ann/.
Sutter, H. and Larus, J. (2005). Software and the concur-
rency revolution. ACM Queue, 3(7):54–62.
Vera, G., Jansen, R., and Suppi, R. (2008). R/parallel -
speeding up bioinformatics analysis with R. BMC
Bioinformatics, 9(1):390.
Yu, H. (2009). Rmpi: Interface (wrapper) to MPI
(message-passing interface). http://www.stats.uwo.ca
/faculty/yu/Rmpi.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
256
