
THE PLASMODIUM GLUTATHIONE S-TRANSFERASE
Bioinformatics Characterization and Classification into the Sigma Class
Emilee E. Colón-Lorenzo, Adelfa E. Serrano
Department of Microbiology and Medical Zoology, University of Puerto Rico School of Medicine, San Juan, Puerto Rico
Hugh B. Nicholas Jr, Troy Wymore, Alexander J. Ropelewski
Pittsburgh Supercomputing Center, Pittsburgh, PA, U.S.A.
Ricardo González-Méndez
Department of Radiological Sciences, University of Puerto Rico School of Medicine, San Juan, Puerto Rico
Keywords: Malaria, Plasmodium, Glutathione S-transferase, Bioinformatics analysis, Structural alignment, GST
classification, Structural modeling.
Abstract: Malaria is a global health problem caused by Plasmodium parasites. Glutathione S-transferase (GST) is
involved in the conjugation of glutathione to drugs and toxic compounds. It is postulated that GST plays an
important role in the development of drug resistance. The three-dimensional (3D) structure of Plasmodium
falciparum GST (PfGST) has been solved and previous work indicates that the PfGST cannot be assigned to
any of the known GST classes. We performed sequence analyses, structural modeling and alignment of
GSTs from Plasmodium to known structures of the GST from other organisms to classify PfGST into a GST
family. Sequence alignments using ClustalW, motif analysis using MEME, and phylogenetic analysis using
MEGA4, of Plasmodium GSTs and 38 other GST sequences were done. The alignments and motifs show a
close relationship to the alpha and sigma class of GSTs. The phylogenetic analysis places the Plasmodium
GSTs in the sigma class. A comparison of PfGST with known structures of GSTs reveals high structural
similarity to the sigma class GST, in particular within the H-site and C-terminus of the protein. These
findings allow PfGST to be classified into the sigma class GSTs. These data may open new avenues for the
development of novel antimalarials.
1 INTRODUCTION
Malaria is one of the most devastating diseases in
the world caused by parasites of the genus
Plasmodium. It is estimated that more than 350
million cases of malaria infections and, over one
million deaths occur annually (WHO, 2005). The
inappropriate and indiscriminate use of drugs has led
to the development of drug resistance in parasites
(Russell, 2004; Whitty et al., 2002; Wongsrichanalai
et al., 2002). Efficient and cost effective alternatives
to presently used drugs are not yet available. A
comprehensive understanding of parasite
development and drug resistance will enable the
development of more effective drug therapies to
combat this disease.
Glutathione S-transferases (GSTs) are a family
of detoxification enzymes found in most organisms
that conjugate reduced glutathione (GSH) with toxic
electrophilic organic compounds and drugs. GSTs
have been subdivided into different classes based on
their primary structure, immunological properties
and substrate specificities (Winayanuwattikun and
Ketterman, 2005). The GST classes are widespread
and are present in a variety of organisms. These
classes include the following: alpha, sigma, mu, pi,
theta, zeta and omega classes (Torres-Rivera and
Landa, 2008). Additionally, there are organism-
specific classes which include several GSTs which
are found only in certain kingdoms or phyla:
lambda, phi, and tau in plants; delta, epsilon in
173
E. Colón-Lorenzo E., E. Serrano A., B. Nicholas Jr H., Wymore T., J. Ropelewski A. and González-Méndez R. (2010).
THE PLASMODIUM GLUTATHIONE S-TRANSFERASE - Bioinformatics Characterization and Classification into the Sigma Class.
In Proceedings of the First International Conference on Bioinformatics, pages 173-180
DOI: 10.5220/0002747401730180
Copyright
c
SciTePress

insects; and beta in prokaryotes. Furthermore, the
kappa class is a unique mitochondrial GST.
Altered GSH mediated detoxification is one of
the proposed underlying mechanisms for the
development of malaria drug resistance. A marked
increased in both GSH levels and GSH-related
enzymatic activity was reported in P. berghei and P.
falciparum lines resistant to chloroquine (Dubois et
al., 1995; Meierjohann et al., 2002; Srivastava et al.,
1999b). Recent results showed that the glutathione
biosynthesis is essential for the parasite’s life cycle
and mosquito transmission (Vega-Rodríguez et al.,
2009). These are some of the reasons why GST is
considered a very promising drug target for the
design of antimalarial drugs.
The genomes of P. falciparum (Pf), P. vivax
(Pv), P. knowlesi (Pk) and P. y. yoelii (Py) have
been sequenced (Carlton et al., 2008) and revealed
that the parasite harbors only one GST. In addition,
the three-dimensional (3D) structure of PfGST has
been solved (Burmeister et al., 2003; Fritz-Wolf et
al., 2003; Perbandt et al., 2004). Detailed structural
comparison to representative structures of the alpha,
mu, and pi classes indicate that the PfGST cannot be
assigned to any of the above GST classes (Fritz-
Wolf et al., 2003; Deponte and Becker, 2005). It was
found that PfGST adopts the canonical GST fold and
is enzymatically active as a homodimer. All the
known structures of GSTs reveal a similar overall
fold: a homodimer where each monomer contains an
N-terminal α/β-domain with βαβαββα topology and
a C-terminal α-helical domain. The active site of
GST is located between the two domains. The active
site possesses two binding sites: the G-site, which
binds reduced GSH, and the H-site, which can
accommodate a variety of substrates. In PfGST, the
N-terminal domain possesses the G-site similar to
that of the alpha, mu, and pi classes studied. The C-
terminal domain shields the H-site. Also, the H-site
in PfGST differed from the other GSTs studied. But
more importantly, PfGST only has 5 amino acid
residues after alpha helix 8 and cannot form the
required structural elements of the alpha, mu, and pi
classes (Fritz-Wolf
et al., 2003).
Here we report bioinformatics sequence analyses
and structural modeling of GSTs from Plasmodium.
We performed a phylogenetic analysis of
Plasmodium spp. sequences and 38 other GST
sequences. We have also analyzed the 3D structure
of PfGST to classify the protein into a GST family.
Our results using PfGST suggest that the
Plasmodium GSTs are a unique family related to the
sigma class. This work is significant for the future
design of specific inhibitors for Plasmodium GSTs
which may lead to the development of novel
antimalarials.
2 MATERIALS AND METHODS
2.1 Alignment of the Plasmodium GSTs
The GST sequences recovered from Plasmodium
spp. and used for subsequent sequence analysis are
listed in Table 1 in the Appendix. The amino acid
sequences were aligned using ClustalW program
(Higgins et al., 1994) with the default parameters.
The alignment was visualized using GeneDoc
program (Nicholas et al., 1997) and some manual
editing was made to produce the final alignment.
2.2 Classification of P. falciparum GST
by Alignment and Phylogenetic
Analysis
We performed a BLAST search with PfGST
(Q8MU52) as the query sequence, using the
iProClass database (Wu et al., 2003). The BLAST
search was carried out using the default parameters.
We selected five members of each of the GST
classes (alpha, sigma, pi, mu, delta, tau and theta)
except for the zeta class where three sequences were
chosen. Since Plasmodium is an eukaryotic
organism, we focused on sequences from mammals,
plants and insects, excluding prokaryotic GSTs. The
GST sequences used are listed in Table 1 in the
Appendix. The multiple sequence alignment was
performed using ClustalW (Higgins et al., 1994).
The Multiple Entropy for Motif Elicitation (MEME)
program was used to find 20 conserved motifs using
‘zero or more occurrences per sequence’ pattern for
selection of motifs (Bailey et al., 1994). The
alignment and motifs were visualized with GeneDoc
(Nicholas et al., 1997). The multiple sequence
alignment was trimmed manually and then used to
perform the phylogenetic analysis. The phylogenetic
analysis was done using the MEGA4 program
(Tamura et al., 2007) using the neighbor-joining
algorithm and a bootstrapped data set of 100
replicates. The program FigTree
(http://tree.bio.ed.ac.uk/software/figtree) was used to
visualize the consensus tree from the bootstrap
analysis.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
174

2.3 Classification of P. falciparum GST
by Structural Alignment
Structural alignment of P. falciparum GST (1Q4J)
with a representative member of each of the GST
classes - alpha, sigma, pi and mu - was performed
using the MultiSeq feature in VMD (Roberts et al.,
2006, Humphrey et al., 1996). The 3D structures
were obtained from the Protein Data Bank (PDB)
(Berman et al., 2000) and are listed in Table 2 in the
Appendix. The sigma GST from Onchocerca
volvulus (2HNL) and the human alpha GST (1PKZ)
were used to perform the structural alignments with
PfGST (shown in Figure 4). The detailed structural
superpositions were carried out with VMD,
specifically in the N-terminal domain (G-site), C-
terminal domain (H-site), and α-helix at the C-
terminus of the proteins. Close-up views of the H-
site, G-site and α-helix in the C-terminus were done
to facilitate the analysis of the 3D structure of the
proteins.
3 RESULTS AND DISCUSSION
3.1 Alignment of the Plasmodium GSTs
The multiple sequence alignment of sequences from
Plasmodium spp. GST is shown in Figure 1. The
sequences used in the alignment are listed in Table 1
in the Appendix. The alignment of Plasmodium spp.
GSTs revealed a significant degree of sequence
identity ranging from 80 to 87%.
Figure 1: Multiple sequence alignment of the Plasmodium
GSTs. Amino acids with a 100% identity are shaded in
red, 99-80% identity are in blue and 79-60% identity in
green.
The GST sequences are highly conserved in all four
species of Plasmodium.
3.2 Classification of P. falciparum GST
by Alignment and Phylogenetic
Analysis
A BLAST search in iProClass was performed using
GST from P. falciparum (Q8MU52). We selected
five sequences of each GST classes except for zeta
class where we selected only three sequences. A
total of 38 GST sequences were obtained and are
listed in Table 1 in the Appendix. The multiple
sequence alignment was done using the program
ClustalW and the MEME program was used to find
20 conserved motifs (Figure 2). The multiple
sequence alignment and motif analyses show that the
Plasmodium GSTs appear to be highly related to the
alpha and sigma families of GST. The alignment
was trimmed manually and was used to perform the
phylogenetic analysis (Figure 3). The phylogenetic
tree indicates that Plasmodium spp. GSTs analyzed
are members of the sigma class of GSTs.
3.3 Classification of P. falciparum GST
by Structural Alignment
The three-dimensional structure superimposition
makes possible the classification of P. falciparum
glutathione S-transferase in a specific GST class.
The three-dimensional structures of GST enzymes
from various classes (alpha, sigma, mu and pi) were
compared to the PfGST 3D structure (1Q4J) by
structural alignment using the MultiSeq feature in
the VMD program. The 3D structures used in the
structural alignments are listed in Table 2 in the
Appendix. Structural alignments of PfGST 3D
structure (1Q4J) with alpha and sigma 3D structures
are presented in Figure 4.
Figure 4A shows the structural alignment of PfGST
(1Q4J) with sigma GST from Oncocherca volvulus
(2HNL). A close-up view showing the α-helix of the
C-terminus is represented in Figure 4B. Figure 4C
shows the structural alignment of PfGST (1Q4J)
with the alpha GST from human (1PKZ). A close-up
view showing the C-terminus is represented in
Figure 4D. Analysis of the 3D structural alignment
of PfGST with the sigma class GST structure shows
high structural similarity in the C-terminus (Figure
4B). The 3D structural alignment of PfGST and the
alpha GST demonstrates a good alignment, but the
C-terminus shows low similarity (Figure 4C). In
addition, we can see that PfGST does not have the
extended helix in the C-terminus that is distinctive
of the alpha class of GST (Figure 4D). Structural
alignments of PfGST 3D structure (1Q4J) with mu
and pi 3D structures were performed showing a low
THE PLASMODIUM GLUTATHIONE S-TRANSFERASE - Bioinformatics Characterization and Classification into the
Sigma Class
175
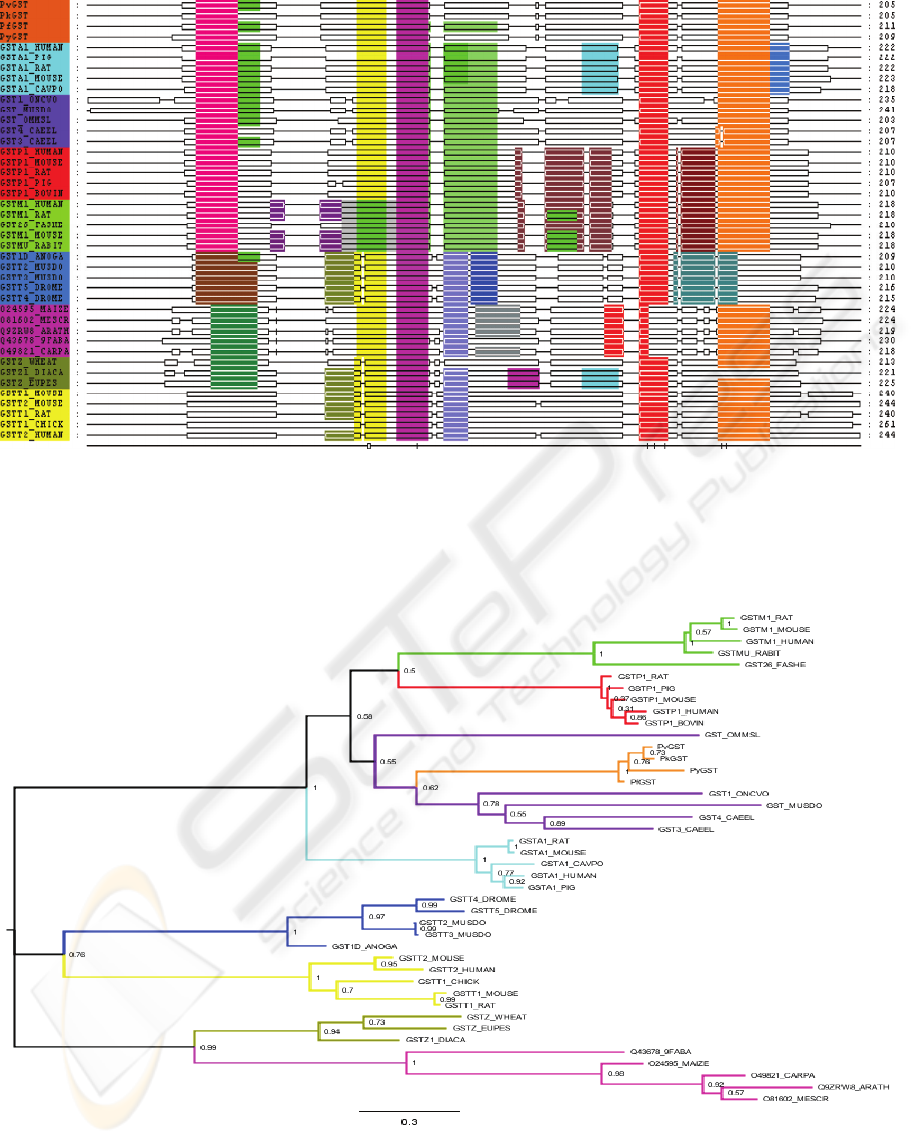
Figure 2: Schematic sequence alignment of four Plasmodium GSTs and 38 GST sequences from eight known GST classes.
Double lines represent where amino acids are present and single lines represent gaps in the alignment. MEME motifs are
identified by different colors and adjusting edges of MEME motifs was the major alignment adjustment. Plasmodium
sequences are colored in orange, alpha sequences in cyan, sigma sequences in purple, pi sequences in red, mu sequences in
green, delta sequences in blue, tau sequences in magenta, zeta sequences in olive green and theta sequences in yellow on the
left side of the figure.
Figure 3: Phylogenetic tree for GST sequences with the fraction of the bootstrap. Plasmodium (orange), sigma (purple),
alpha (cyan), pi (red), mu (green), delta (blue), tau (magenta), zeta (olive green) and theta (yellow).
BIOINFORMATICS 2010 - International Conference on Bioinformatics
176
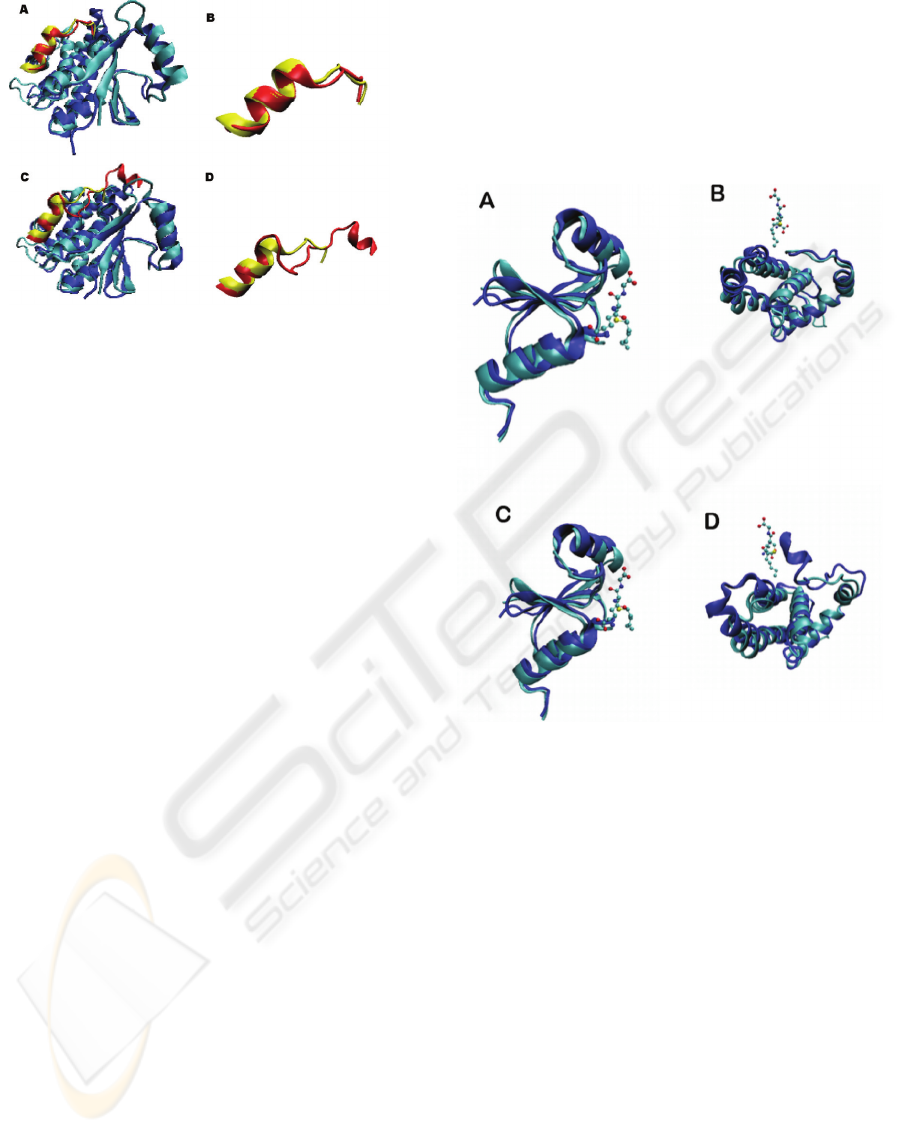
Figure 4: Structural alignment of P. falciparum GST with
sigma and alpha class GST structure. (A) Structural
alignment of PfGST (cyan) and a sigma class GST (blue);
(B) Close-up view showing the C-terminus of PfGST
(yellow) and sigma GST (red); (C) Structural alignment of
PfGST (cyan) and an alpha class GST (blue); (D) Close-
up view showing the C-terminus of PfGST (yellow) and
alpha GST (red).
structural similarity (data not shown). An important
feature of the C-terminus of alpha class GST is an
alpha helix close to the active site. This helix is an
essential element of the GST alpha class (Nilsson et
al., 2002). Based on this, we performed an analysis
of this area of the protein using structural
alignments. Fritz-Wolf et al. (2003) described the
PfGST having a different length C-terminus - only
has 5 amino acid residues after alpha helix 8 - and
therefore could not form the required structural
elements from mammalian GSTs as part of what
makes them not classifiable. Our observation is that
PfGST aligned well with the sigma class GST from
O. volvulus. We find that using common structural
features and MEME patterns GSTs have a very
consistent C-terminus length within each class and
that is one of the features that helped clarify the
classification of the Plasmodium sequences as a
sigma class in both the structural alignment (Figure
4) and the sequence alignment (Figure 2).
Furthermore, analysis of the 3D structure of
PfGST in the N-terminal domain (G-site) and C-
terminal domain (H-site) was performed. Structural
alignment of the N-terminal domain, that contains
the G-site, reveals that PfGST shares a common
backbone fold with the sigma (2HNL) GST from O.
volvulus and alpha (1PKZ) GST from human. Both
structure alignments show a similar binding mode
for S-hexylglutathione (GSH derivative). Close
views of the N-terminal domain are shown (Figures
5A and 5C, respectively). Structural alignment of the
C-terminal domain, that contains the H-site, shows
that PfGST shares a common backbone with sigma
(2HNL) GST, while alpha (1PKZ) GST is different,
specifically in the C-terminus (Figures 5B and 5D,
respectively). Generally, the H-site of GST enzymes
is more variable than the G-site due to the great
number of secondary structures (Fritz-Wolf et al.,
2003). These findings are in agreement with Fritz-
Wolf et al. (2003).
Figure 5: Structural comparison of GST structures from P.
falciparum GST with a sigma GST from O. volvulus and
an alpha GST from human. (A) Structural alignment of N-
terminal (G-site) of PfGST (cyan) and a sigma class GST
(blue); (B) Structural alignment of C-terminal (H-site) of
PfGST (cyan) and a sigma class GST (blue); (C)
Structural alignment of N-terminal (G-site) of PfGST
(cyan) and a alpha class GST (blue); (D) Structural
alignment of C-terminal (H-site) of PfGST (cyan) and a
alpha class GST (blue). The S-hexylglutathione ligand is
shown as a ball-and-stick model.
A comprehensive analysis of the 3D structure of
PfGST in the H-site, G-site and C-terminus of the
proteins was done and established that these regions
have high similarity with the sigma class GST from
O. volvulus (2HNL). The structural alignments
support the results obtained with the phylogenetic
tree. These results allow us to classify the
Plasmodium GSTs as members of the sigma class of
GSTs.
THE PLASMODIUM GLUTATHIONE S-TRANSFERASE - Bioinformatics Characterization and Classification into the
Sigma Class
177

4 CONCLUSIONS
The GST sequences of four Plasmodium species
were compared using multiple sequence alignments
and found to be highly conserved. When aligned
with members of each of the GST classes used in
this study our results show that the Plasmodium
GSTs appear to be highly related to the alpha and
sigma families of GSTs. Using phylogenetic analysis
we found that Plasmodium GST are members of the
sigma class. The 3D structure of PfGST was
compared, using structural alignments, with
members of several GST classes. We determined
that PfGST has a high degree of similarity with the
sigma class in the H-site, G-site and C-terminus of
the protein. Structural analysis and phylogenetic
analysis of PfGST revealed that this enzyme
represents a unique clade within the sigma class of
GSTs.
This work contributes to a better understanding
of the structure and classification of Plasmodium
GSTs. These results have the potential to enhance
our knowledge of the relevance of GST to
Plasmodium drug resistance. It has been recently
demonstrated that 7-nitro-2,1,3-benzoxadiazole
derivatives are a new class of suicide inhibitors of
GST that accumulate in tumor cells and evade the
extrusion mechanisms mediated by the multidrug
resistance associated protein pumps (MRP) (Ricci et
al., 2005). Studies in human cancer cells in-vitro
showed that the GST inhibitor 6-(7-nitro-2,1,3-
benzoxadiazol-4-ylthio)hexanol (NBDHEX) has
high antiproliferative activity and helped overcome
MRP mediated drug resistance (Federici et el., 2009;
Filomeni et al., 2008). In-silico studies will be
undertaken to evaluate NBDHEX’s potential as an
antimalarial drug. These studies could support the
use of this GST inhibitor in Plasmodium.
We expect to use the results presented here for
further studies using molecular biology and genetics
approaches in order to study the involvement of
GST in the Plasmodium life cycle and mosquito
transmission. These studies should be useful to find
alternative strategies for malaria control.
ACKNOWLEDGEMENTS
RGM, EECL, HBN, TW, AJP and this project were
partially supported by NIGMS grant T36-
GM008789 and acknowledge use of the Pittsburgh
Supercomputing Center’s National Resource for
Biomedical Supercomputing resources funded
through NIH NCRR grant P41-RR06009. AES and
EECL were partially supported by NIGMS/MBRS
award GM08224 and from the RCMI award
G12RR03051 of the NCRR/NIH.
REFERENCES
Bailey, T.L., Elkan, C., 1994. "Fitting a mixture model by
expectation maximization to discover motifs in
biopolymers", Proceedings of the Second International
Conference on Intelligent Systems for Molecular
Biology, pp. 28-36, AAAI Press, Menlo Park,
California.
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G.,
Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne,
P.E., 2000. The Protein Data Bank Nucleic Acids
Research, 28 pp. 235-242.
Burmeister, C., Perbandt, M., Betzel, Ch., Walter, R. D.,
Liebau, E., 2003. Crystallization and preliminary X-
ray diffraction studies of the glutathione S-transferase
from Plasmodium falciparum. Acta Crystallogr. D
Biol. Crystallogr. 59, 1469–1471.
Carlton, J.M., Adams, J.H., Silva, J.C., et al., 2008.
Comparative genomics of the neglected human
malaria parasite Plasmodium vivax. Nature 455:757-
763.
Deponte, M., Becker, K., 2005. Glutathione S-transferase
from malarial parasites: structural and functional
aspects. Methods Enzymol. 401:240–252.
Dubois, V.L., Platel, D.F., Pauly, G., and Tribouley-Duret,
J. 1995. Plasmodium berghei: Implication of
intracellular glutathione and its related enzyme in
chloroquine resistance in vivo. Exp. Parasitol. 81: 117-
124.
Federici, L., Sterzo, C.L., Pezzola, S., Matteo, A.D.,
Scaloni, F., Federici, G., Caccuri, A.M. 2009.
Structural basis for the binding of the anticancer
compound 6-(7-Nitro-2,1,3-Benzoxadiazol-4-
Ylthio)Hexanol to human glutathione S-transferases.
Cancer Research 69:20.
Filomeni G., Turella, P., Dupuis, M.L., Forini, O., Ciriolo,
M.R., Cianfriglia, M., Pezzola, S., Federici, G.,
Caccuri, A.M. 2008. 6-(7-Nitro-2,1,3-Benzoxadiazol-
4-Ylthio)Hexanol, a specific glutathione S-transferase
inhibitor, overcomes the multidrug resistance (MDR)-
associated protein 1-mediated MDR in small cell lung
cancer. Molecular cancer therapeutics 7(2).
Fritz-Wolf, K., Becker, A., Rahlfs, S., Harwaldt, P.,
Schirmer, R. H., Kabsch, W., Becker, K., 2003. X-ray
structure of glutathione S-transferase from the malarial
parasite Plasmodium falciparum. Proc. Natl. Acad.
Sci. USA 100, 13821–13826.
Higgins, D., Thompson, J., Gibson, T., Thompson, J.D.,
Higgins, D.G., Gibson, T.J., 1994. CLUSTAL W:
improving the sensitivity of progressive multiple
sequence alignment through sequence weighting,
position-specific gap penalties and weight matrix
choice. Nucleic Acids Res 22:4673-4680.
BIOINFORMATICS 2010 - International Conference on Bioinformatics
178

Humphrey, W., Dalke, A., Schulten, K., 1996. VMD -
Visual Molecular Dynamics. J. Molec. Graphics, vol.
14, pp. 33-38
Meierjohann, S., Walter, R.D., Muller, S. 2002.
Regulation of intracellular glutathione levels in
erythrocytes infected with chloroquine-sensitive and
chloroquine-resistant Plasmodium falciparum.
Biochem J. Dec 15: 368, 761-8.
Nicholas, K.B., Nicholas, H.B. Jr,, Deerfield, D.W. II,
1997. GeneDoc: Analysis and Visualization of Genetic
Variation. EMBNEW NEWS 4:14.
Nilsson, L. O., Edalat, M., Pettersson, P. L., Mannervik,
B., 2002. Aromatic residues in the C-terminal region
of glutathione transferase A1-1 influence rate-
determining steps in the catalytic mechanism.
Biochimica et Biophysica Acta 29: 1598(1-2):199-
205.
Perbandt, M., Burmeister, C., Walter, R. D., Betzel, C.,
Liebau, E., 2004. Native and inhibited structure of a
Mu class-related glutathione S-transferase from
Plasmodium falciparum. J. Biol. Chem. 279, 1336–
1342.
Rambaut, A., 2006. FigTree: Tree figure drawing tool,
version 1.0. Available from http://tree.bio.
bio.ed.ac.uk/software/figtree/, Institute of
Evolutionary Biology, University of Edinburgh.
Ricci, G., De Maria, F., Antonini, G., Turella, P., Bullo,
A., Stella, L., Filomeni, G., Federici, G., Caccuri,
A.M. 2005. 7-Nitro-2,1,3-benzoxadiazole derivatives,
a new class of suicide inhibitors for glutathione S-
transferases. The Journal of Biological Chemistry
280:28, 26397-26405.
Roberts, E., Eargle, J., Wright, D., Luthey-Schulten, Z.,
2006. MultiSeq: unifying sequence and structure data
for evolutionary analysis. BMC Bioinformatics. Aug
16;7:382.
Rusell, S., 2004. The economic burden of illness for
households in developing countries: a review of
studies focusing on malaria, tuberculosis, and human
immunodeficiency virus/acquired immunodeficiency
syndrome. Am J Trop Med Hyg.71 (2 Suppl):147-55.
Srivastava, P., Puri, S.K., Kamboj, K.K., and Pandey, V.C.
1999. Glutathione S-transferase activity in malarial
parasites. Trop. Med. Int. Health 4:251-254.
Tamura, K., Dudley, J., Nei, M., Kumar, S., 2007.
MEGA4: Molecular Evolutionary Genetics Analysis
(MEGA) software version 4.0. Molecular Biology and
Evolution 24: 1596-1599.
Torres-Rivera, A., Landa, A., 2008. Glutathione
transferases from parasites: A biochemical view. Acta
Tropica 105: 99–112.
Vega-Rodríguez, J., Franke-Fayard, B., Dinglasan, R.R.,
Janse, C.J., Pastrana-Mena, R., Waters, A.P., Coppens,
I., Rodríguez-Orengo, J.F., Jacobs-Lorena, M.,
Serrano, A.E. 2009. The glutathione biosynthetic
pathway of Plasmodium is essential for mosquito
transmission. PLoS Pathogens 5:2.
Winayanuwattikun, P., Ketterman, A.J., 2005. An
electron-sharing network involved in the catalytic
mechanism is functionally conserved in different
glutathione transferase classes. The Journal of
Biological Chemistry.
Whitty, C.J., Rowland, M., Sanderson, F., Mutabingwa,
T.K., 2002. Science, medicicne, and the future:
Malaria. BMJ 325(7374):1221-1224.
WHO, 2005. Global malaria situation. http://www.who.
int/globalatlas/autologin/malria_login.asp
Wongsrichanalai, C., Pickard, A.L., Wernsdorfer, W.H.,
Meshnick, S.R. 2002. Epidemiology of drug-resistant
malaria. Lancet Infect Dis. 4:209-18.
Wu, C.H., Huang, H., Nikolskaya, A., Hu, Z., Barker,
W.C., 2004. The iProClass integrated database for
protein functional analysis. Comput Biol Chem.
Feb;28(1):87-96.
THE PLASMODIUM GLUTATHIONE S-TRANSFERASE - Bioinformatics Characterization and Classification into the
Sigma Class
179
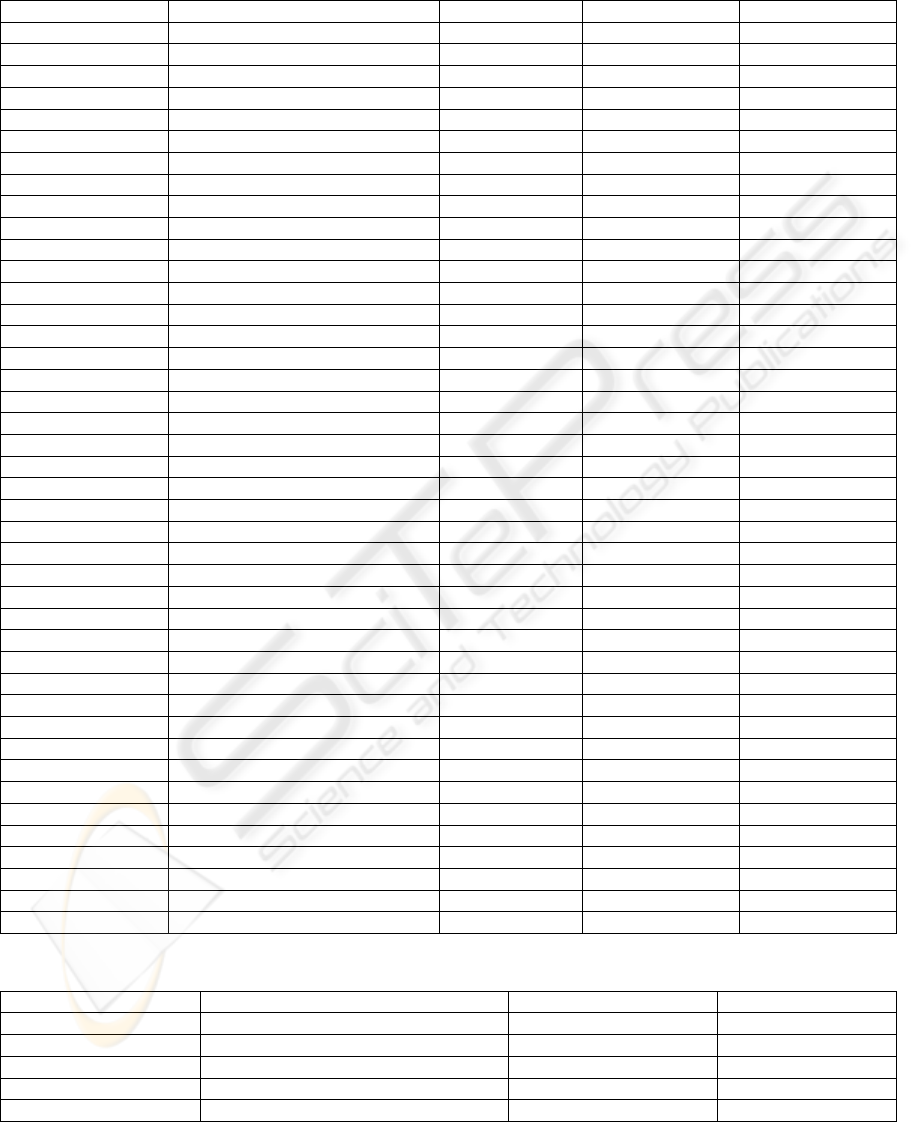
APPENDIX
Table 1: Glutathione S-transferase sequences used in the alignments.
Protein name Organism Family class Accession number Protein length (aa)
PfGST Plasmodium falciparum sigma Q8MU52 211
PvGST Plasmodium vivax sigma Q0ZS46 205
PkGST Plasmodium knowlesi sigma B3LAI5 205
PyGST Plasmodium yoelii sigma Q7REH6 209
GSTA1_HUMAN Homo sapiens alpha P08263 222
GSTA1_RAT Rattus norvegicus alpha P00502 222
GSTA1_MOUSE Mus musculus alpha P13745 223
GSTA1_PIG Sus scrofa alpha P51781 222
GSTA1_CAVPO Cavia porcellus alpha P81706 218
GST1_ONCVO Oncocherca volvulus sigma P46434 235
GST_MUSDO Musca domestica sigma P46437 241
GST_OMMSL Ommastrephes sloanei sigma P46088 203
GST4_CAEEL Caenorhabditis elegans sigma Q21355 207
GST3_CAEEL Caenorhabditis elegans sigma O16116 207
GSTP1_HUMAN Homo sapiens pi P09211 210
GSTP1_MOUSE Mus musculus pi P19157 210
GSTP1_RAT Rattus norvegicus pi P04906 210
GSTP1_PIG Sus scrofa pi P80031 207
GSTP1_BOVIN Bos taurus pi P28801 210
GSTM1_HUMAN Homo sapiens mu P09488 218
GSTM1_RAT Rattus norvegicus mu P04905 218
GST26_FASHE Fasciola hepatica mu P30112 218
GSTM1_MOUSE Mus musculus mu P10649 218
GSTMU_RABIT Oryctolagus cuniculus mu P46409 218
GST1D_ANOGA Anopheles gambiae delta Q93113 209
GSTT2_MUSDO Musca domestica delta P46431 210
GSTT3_MUSDO Musca domestica delta P46432 210
GSTT5_DROME Drosophila melanogaster delta Q9VG95 216
GSTT4_DROME Drosophila melanogaster delta Q9VG96 215
O24595_MAIZE Zea mays tau O24595 224
O81602_MESCR Mesembryanthemum crystallinum tau O81602 224
Q9ZRW8_ARATH Arabidopsis thaliana tau Q9ZRW8 219
Q43678_9FABA Vigna radiate tau Q43678 230
O49821_CARPA Carica papaya tau O49821 218
GSTZ-WHEAT Triticum aestivum zeta O04437 213
GSTZ1_DIACA Dianthus caryophyllus zeta P28342 221
GSTZ_EUPES Euphorbia esula zeta P57108 225
GSTT1_MOUSE Mus musculus theta Q64471 240
GSTT2_MOUSE Mus musculus theta Q61133 244
GSTT1_RAT Rattus norvegicus theta Q01579 240
GSTT1_CHICK Gallus gallus theta P20135 261
GSTT2_HUMAN Homo sapiens theta P30712 244
Table 2: Glutathione S-transferase sequences used for the structural alignments.
Protein name Organism Family class PDB code
PfGST Plasmodium falciparum sigma 1Q4J
GST1_ONCVO Oncoherca volvulus sigma 2HNL
DmGST Drosophila melanogaster sigma 1MOU
GSTA1_HUMAN Homo sapiens alpha 1PKZ
GSTM1_HUMAN Homo sapiens mu 1GTU
BIOINFORMATICS 2010 - International Conference on Bioinformatics
180
